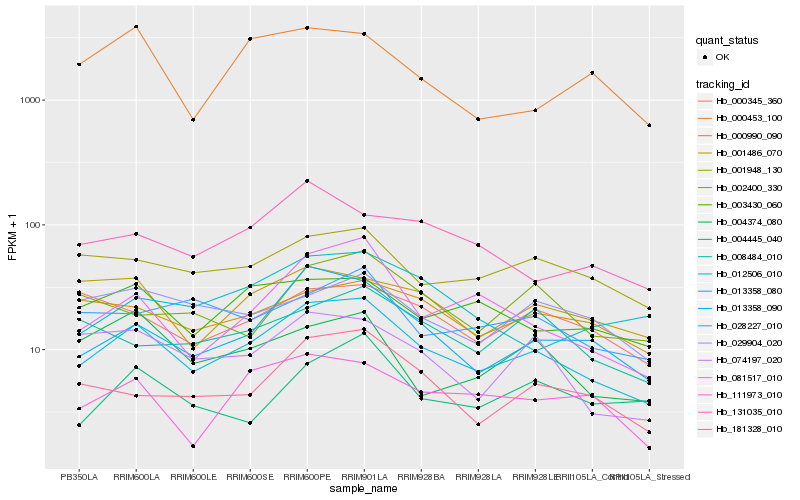
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013358_080 |
0.0 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 2 |
Hb_013358_090 |
0.1093322892 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 3 |
Hb_081517_010 |
0.1345711616 |
- |
- |
PREDICTED: putative potassium transporter 12 isoform X2 [Jatropha curcas] |
| 4 |
Hb_181328_010 |
0.1367161082 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_001948_130 |
0.1438403827 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 6 |
Hb_111973_010 |
0.144277528 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_074197_020 |
0.145947761 |
- |
- |
PREDICTED: uncharacterized protein LOC105645948 [Jatropha curcas] |
| 8 |
Hb_003430_060 |
0.1475502725 |
- |
- |
PREDICTED: potassium transporter 23 isoform X2 [Elaeis guineensis] |
| 9 |
Hb_000990_090 |
0.1529413538 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_001486_070 |
0.1530708222 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 11 |
Hb_131035_010 |
0.1531802712 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_012506_010 |
0.1533637283 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 13 |
Hb_000453_100 |
0.1595022912 |
- |
- |
PREDICTED: metallothionein-like protein 1 [Sesamum indicum] |
| 14 |
Hb_004374_080 |
0.1596923521 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 15 |
Hb_028227_010 |
0.1602988787 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 16 |
Hb_000345_360 |
0.1607328684 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 17 |
Hb_002400_330 |
0.1615532715 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_029904_020 |
0.1617002508 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 19 |
Hb_004445_040 |
0.1624021084 |
- |
- |
PREDICTED: uncharacterized protein LOC105639134 isoform X5 [Jatropha curcas] |
| 20 |
Hb_008484_010 |
0.1631308497 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |