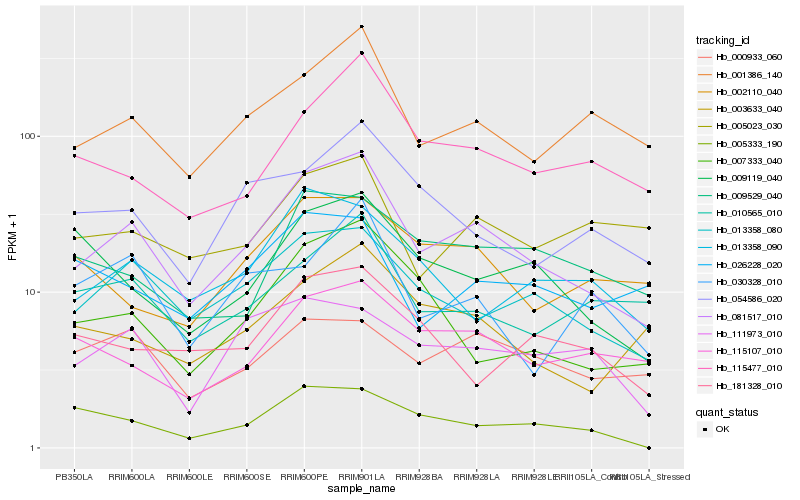
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_081517_010 |
0.0 |
- |
- |
PREDICTED: putative potassium transporter 12 isoform X2 [Jatropha curcas] |
| 2 |
Hb_013358_080 |
0.1345711616 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 3 |
Hb_001386_140 |
0.1493226779 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 4 |
Hb_115477_010 |
0.1649472814 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 5 |
Hb_007333_040 |
0.1664118571 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 6 |
Hb_115107_010 |
0.1702263475 |
- |
- |
Early-responsive to dehydration stress protein (ERD4) isoform 1 [Theobroma cacao] |
| 7 |
Hb_003633_040 |
0.1703031482 |
- |
- |
hypothetical protein B456_008G146200 [Gossypium raimondii] |
| 8 |
Hb_054586_020 |
0.171154519 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 9 |
Hb_013358_090 |
0.1724190337 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 10 |
Hb_010565_010 |
0.1751047108 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 11 |
Hb_009119_040 |
0.1786365265 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2A [Jatropha curcas] |
| 12 |
Hb_009529_040 |
0.1819992515 |
- |
- |
PREDICTED: transmembrane protein 120 homolog isoform X1 [Malus domestica] |
| 13 |
Hb_111973_010 |
0.1822046123 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 14 |
Hb_002110_040 |
0.1854609479 |
- |
- |
hypothetical protein AMTR_s00135p00074190 [Amborella trichopoda] |
| 15 |
Hb_005023_030 |
0.1857814505 |
- |
- |
PREDICTED: uncharacterized protein LOC103417254 [Malus domestica] |
| 16 |
Hb_000933_060 |
0.1867631247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005333_190 |
0.1870588752 |
- |
- |
PREDICTED: AP-4 complex subunit mu isoform X2 [Populus euphratica] |
| 18 |
Hb_030328_010 |
0.1881334211 |
- |
- |
hypothetical protein CICLE_v10015936mg [Citrus clementina] |
| 19 |
Hb_026228_020 |
0.1885083023 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 20 |
Hb_181328_010 |
0.1891586677 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |