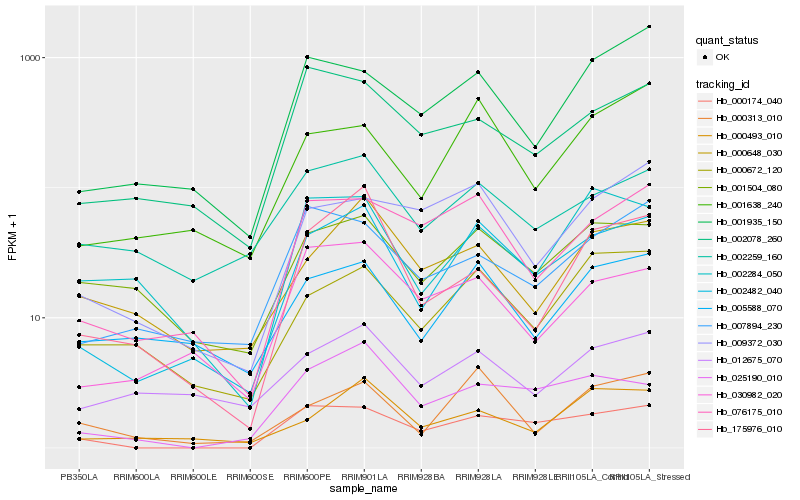
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002482_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 2 |
Hb_076175_010 |
0.1478340815 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB7 [Cucumis melo] |
| 3 |
Hb_001638_240 |
0.1517335925 |
- |
- |
PREDICTED: uncharacterized protein LOC100262493 [Vitis vinifera] |
| 4 |
Hb_005588_070 |
0.1561896602 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 5 |
Hb_000672_120 |
0.1591685787 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 6 |
Hb_030982_020 |
0.1616851474 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_000174_040 |
0.1620332797 |
- |
- |
- |
| 8 |
Hb_025190_010 |
0.1663465917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001504_080 |
0.1670980191 |
- |
- |
golgi snare 11 protein, putative [Ricinus communis] |
| 10 |
Hb_000493_010 |
0.1684409846 |
- |
- |
hypothetical protein CICLE_v10021431mg [Citrus clementina] |
| 11 |
Hb_000313_010 |
0.1692480496 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 12 |
Hb_001935_150 |
0.1728327905 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 13 |
Hb_002259_160 |
0.1741398045 |
- |
- |
hypothetical protein PSEUBRA_SCAF14g01508 [Pseudozyma brasiliensis GHG001] |
| 14 |
Hb_007894_230 |
0.1766626587 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 15 |
Hb_012675_070 |
0.1783552893 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Pyrus x bretschneideri] |
| 16 |
Hb_000648_030 |
0.1789316002 |
- |
- |
hypothetical protein PHAVU_002G055100g [Phaseolus vulgaris] |
| 17 |
Hb_002078_260 |
0.1796907713 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_009372_030 |
0.1801533263 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |
| 19 |
Hb_002284_050 |
0.1812842293 |
- |
- |
hypothetical protein POPTR_0002s00740g [Populus trichocarpa] |
| 20 |
Hb_175976_010 |
0.1815990521 |
- |
- |
- |