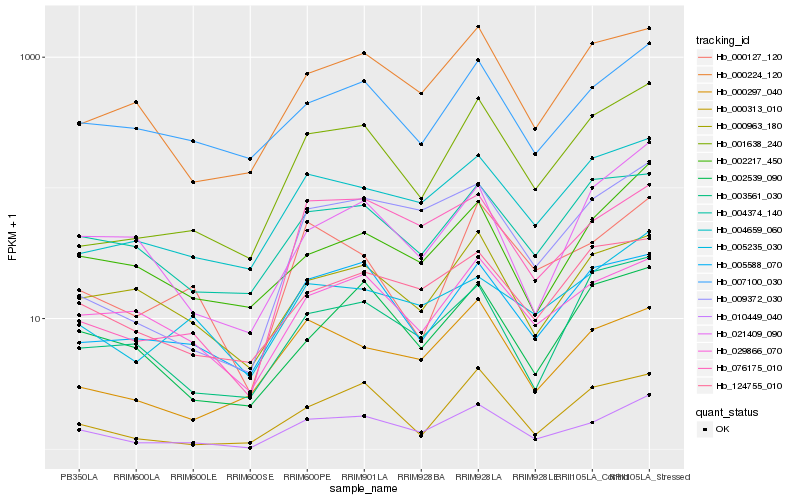
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010449_040 |
0.0 |
- |
- |
PREDICTED: probable CDP-diacylglycerol--inositol 3-phosphatidyltransferase 2 [Jatropha curcas] |
| 2 |
Hb_009372_030 |
0.1383289726 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |
| 3 |
Hb_001638_240 |
0.145282229 |
- |
- |
PREDICTED: uncharacterized protein LOC100262493 [Vitis vinifera] |
| 4 |
Hb_000224_120 |
0.1469476666 |
- |
- |
eukaryotic translation initiation factor 1Aa [Hevea brasiliensis] |
| 5 |
Hb_007100_030 |
0.1487788586 |
- |
- |
40S ribosomal protein S17B [Hevea brasiliensis] |
| 6 |
Hb_029866_070 |
0.1518969621 |
- |
- |
PREDICTED: myosin IB heavy chain [Jatropha curcas] |
| 7 |
Hb_005588_070 |
0.1526933692 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 8 |
Hb_124755_010 |
0.1534986187 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 9 |
Hb_000297_040 |
0.1539342808 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 10 |
Hb_002539_090 |
0.154675453 |
- |
- |
PREDICTED: uncharacterized protein LOC103489178 [Cucumis melo] |
| 11 |
Hb_076175_010 |
0.154758835 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB7 [Cucumis melo] |
| 12 |
Hb_000313_010 |
0.1552996626 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_004659_060 |
0.1558245406 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 14 |
Hb_002217_450 |
0.1559783322 |
- |
- |
PREDICTED: probable prefoldin subunit 3 [Jatropha curcas] |
| 15 |
Hb_021409_090 |
0.157580716 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0003s14450g [Populus trichocarpa] |
| 16 |
Hb_005235_030 |
0.1578158382 |
- |
- |
PREDICTED: dolichol phosphate-mannose biosynthesis regulatory protein [Eucalyptus grandis] |
| 17 |
Hb_000963_180 |
0.1578853757 |
- |
- |
calcium/calmodulin-dependent protein kinase kinase, putative [Ricinus communis] |
| 18 |
Hb_000127_120 |
0.1599323643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003561_030 |
0.1602603551 |
- |
- |
PREDICTED: uncharacterized protein LOC105645274 [Jatropha curcas] |
| 20 |
Hb_004374_140 |
0.1622660812 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |