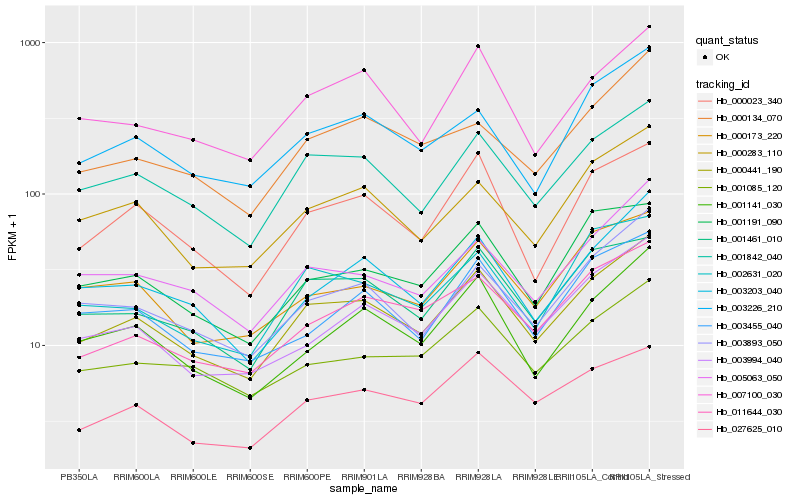
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000441_190 |
0.0 |
- |
- |
PREDICTED: GDP-L-galactose phosphorylase 1-like isoform X2 [Vitis vinifera] |
| 2 |
Hb_001842_040 |
0.0512286545 |
- |
- |
PREDICTED: uncharacterized protein LOC100800807 isoform X2 [Glycine max] |
| 3 |
Hb_003893_050 |
0.0731939809 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000283_110 |
0.073920846 |
- |
- |
PREDICTED: probable prefoldin subunit 4 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001461_010 |
0.0776351447 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 5 [Pyrus x bretschneideri] |
| 6 |
Hb_003994_040 |
0.0777528356 |
- |
- |
- |
| 7 |
Hb_000023_340 |
0.078680291 |
- |
- |
PREDICTED: enhancer of rudimentary homolog [Jatropha curcas] |
| 8 |
Hb_003203_040 |
0.0786884369 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 7 [Vitis vinifera] |
| 9 |
Hb_011644_030 |
0.0786998646 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Pyrus x bretschneideri] |
| 10 |
Hb_001141_030 |
0.0804408209 |
- |
- |
hypothetical protein PRUPE_ppa024854mg [Prunus persica] |
| 11 |
Hb_003226_210 |
0.0806660697 |
- |
- |
hypothetical protein PRUPE_ppa013414mg [Prunus persica] |
| 12 |
Hb_007100_030 |
0.0811252988 |
- |
- |
40S ribosomal protein S17B [Hevea brasiliensis] |
| 13 |
Hb_001085_120 |
0.0819485481 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X1 [Jatropha curcas] |
| 14 |
Hb_005063_050 |
0.0827708835 |
- |
- |
PREDICTED: probable prefoldin subunit 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002631_020 |
0.0842009169 |
- |
- |
PREDICTED: protein FAM32A-like [Jatropha curcas] |
| 16 |
Hb_027625_010 |
0.0850416578 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 17 |
Hb_000173_220 |
0.0861562891 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 18 |
Hb_000134_070 |
0.0872569598 |
- |
- |
60S ribosomal protein L6, putative [Ricinus communis] |
| 19 |
Hb_001191_090 |
0.0878626233 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 20 |
Hb_003455_040 |
0.0881151648 |
- |
- |
hypothetical protein JCGZ_17270 [Jatropha curcas] |