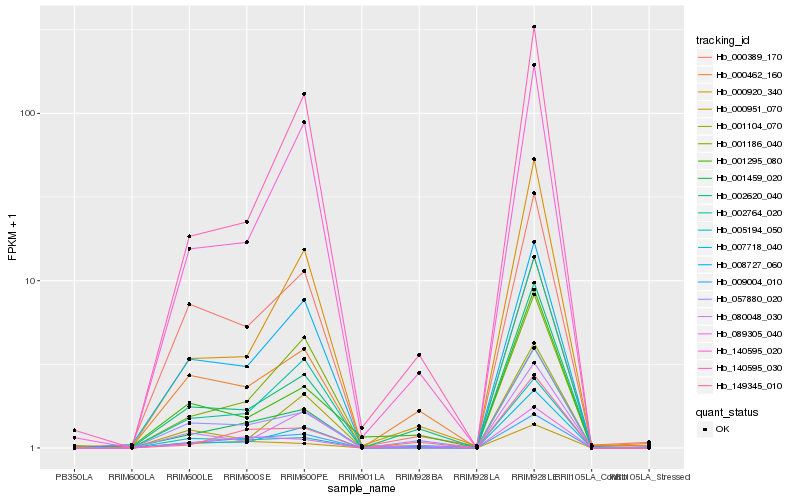
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001459_020 |
0.0 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 2 |
Hb_002620_040 |
0.1045508503 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 3 |
Hb_007718_040 |
0.1059665985 |
- |
- |
hypothetical protein PRUPE_ppb025054mg [Prunus persica] |
| 4 |
Hb_057880_020 |
0.106691444 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 5 |
Hb_089305_040 |
0.1124383829 |
- |
- |
- |
| 6 |
Hb_000920_340 |
0.1228260105 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_149345_010 |
0.125416966 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_008727_060 |
0.1294682068 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 9 |
Hb_000389_170 |
0.1302724817 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 10 |
Hb_000462_160 |
0.1318983463 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_140595_030 |
0.1346841222 |
- |
- |
- |
| 12 |
Hb_002764_020 |
0.1350855447 |
- |
- |
hypothetical protein JCGZ_24084 [Jatropha curcas] |
| 13 |
Hb_140595_020 |
0.1380746425 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Solanum lycopersicum] |
| 14 |
Hb_080048_030 |
0.1399272311 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 15 |
Hb_009004_010 |
0.1404389166 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 16 |
Hb_000951_070 |
0.1412004802 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 17 |
Hb_005194_050 |
0.1441264601 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 18 |
Hb_001295_080 |
0.1457167206 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 19 |
Hb_001186_040 |
0.1462905865 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |
| 20 |
Hb_001104_070 |
0.1472822909 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |