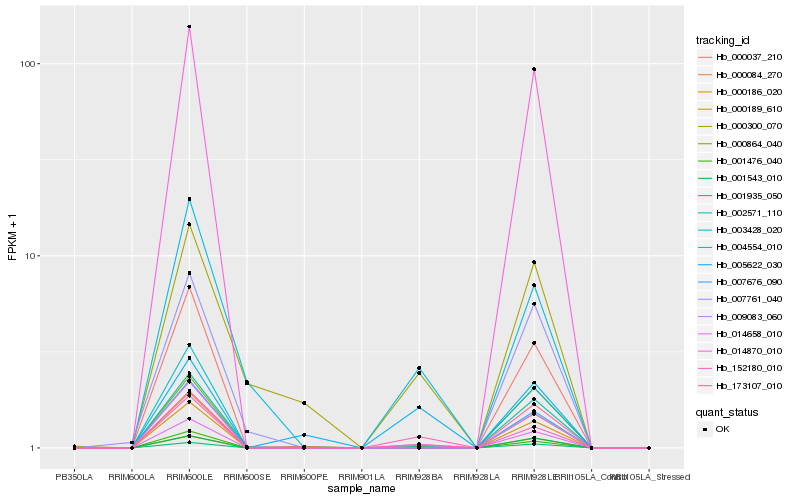
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014658_010 |
0.0 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 2 |
Hb_152180_010 |
0.1075468938 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 3 |
Hb_173107_010 |
0.1315241598 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 4 |
Hb_002571_110 |
0.1579979755 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 5 |
Hb_004554_010 |
0.1742585832 |
- |
- |
PREDICTED: uncharacterized protein LOC105636233 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001476_040 |
0.1774938233 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_000186_020 |
0.1823379238 |
- |
- |
PREDICTED: uncharacterized protein LOC104901327 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_000084_270 |
0.1823815557 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 9 |
Hb_014870_010 |
0.1842920066 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_007676_090 |
0.1851211357 |
- |
- |
PREDICTED: uncharacterized protein LOC105644038 [Jatropha curcas] |
| 11 |
Hb_003428_020 |
0.1859178725 |
- |
- |
PREDICTED: uncharacterized protein LOC105140247 [Populus euphratica] |
| 12 |
Hb_000037_210 |
0.1864366016 |
- |
- |
- |
| 13 |
Hb_009083_060 |
0.1866131441 |
- |
- |
PREDICTED: uncharacterized protein LOC103344557 [Prunus mume] |
| 14 |
Hb_001935_050 |
0.1926089683 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 [Jatropha curcas] |
| 15 |
Hb_000300_070 |
0.1987296157 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_007761_040 |
0.198946577 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 17 |
Hb_000864_040 |
0.1993635429 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000189_610 |
0.2011811456 |
- |
- |
Late embryogenesis abundant protein D-34, putative [Ricinus communis] |
| 19 |
Hb_001543_010 |
0.2016837768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005622_030 |
0.2038619181 |
- |
- |
transferase, putative [Ricinus communis] |