| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
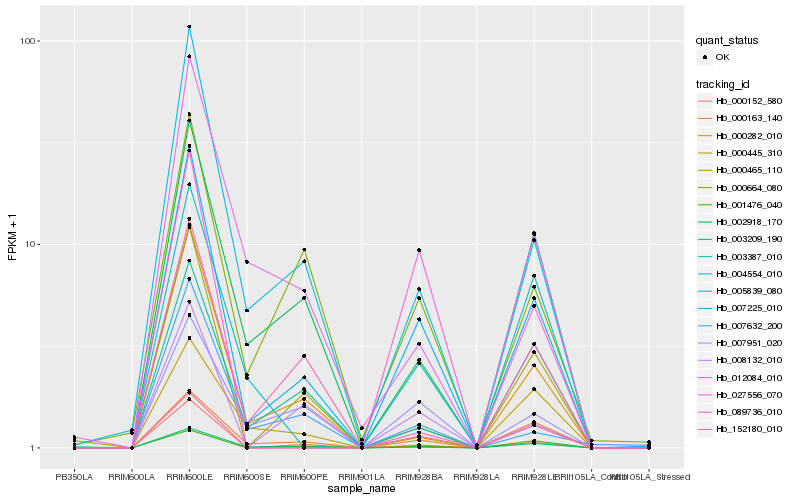
Hb_007225_010 |
0.0 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 2 |
Hb_005839_080 |
0.1398207664 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 3 |
Hb_000152_580 |
0.1430874796 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 4 |
Hb_000163_140 |
0.1675556107 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 5 |
Hb_008132_010 |
0.1718627408 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 6 |
Hb_001476_040 |
0.1741517723 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_002918_170 |
0.1775447701 |
- |
- |
PREDICTED: MACPF domain-containing protein NSL1 [Jatropha curcas] |
| 8 |
Hb_000664_080 |
0.1851533619 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X2 [Jatropha curcas] |
| 9 |
Hb_152180_010 |
0.1904332578 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 10 |
Hb_007951_020 |
0.1911838509 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 11 |
Hb_003387_010 |
0.1924440471 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 12 |
Hb_000465_110 |
0.1930295837 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Jatropha curcas] |
| 13 |
Hb_012084_010 |
0.1934007278 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 14 |
Hb_003209_190 |
0.199749021 |
- |
- |
hypothetical protein POPTR_0010s16770g [Populus trichocarpa] |
| 15 |
Hb_000445_310 |
0.2001604564 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 16 |
Hb_007632_200 |
0.2017441867 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 17 |
Hb_027556_070 |
0.2022429372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004554_010 |
0.2026514025 |
- |
- |
PREDICTED: uncharacterized protein LOC105636233 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000282_010 |
0.2026825308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_089736_010 |
0.2036710138 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |