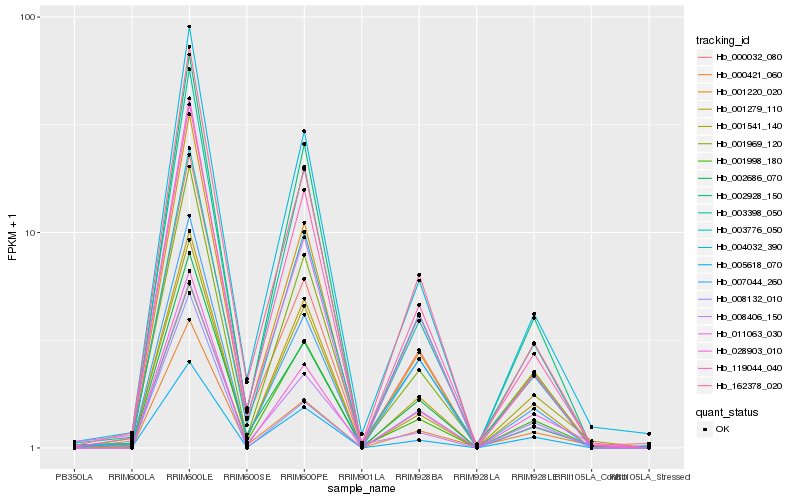
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028903_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_09851 [Jatropha curcas] |
| 2 |
Hb_000421_060 |
0.0664660482 |
- |
- |
PREDICTED: uncharacterized protein LOC105628158 [Jatropha curcas] |
| 3 |
Hb_000032_080 |
0.0931839082 |
transcription factor |
TF Family: HMG |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_008406_150 |
0.0988929258 |
- |
- |
PREDICTED: mitotic checkpoint serine/threonine-protein kinase BUB1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002686_070 |
0.1019101039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005618_070 |
0.1020762751 |
- |
- |
PREDICTED: uncharacterized protein LOC105643318 [Jatropha curcas] |
| 7 |
Hb_001220_020 |
0.1040514029 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 8 |
Hb_001969_120 |
0.1054509268 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 9 |
Hb_162378_020 |
0.1086400324 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 10 |
Hb_004032_390 |
0.10866873 |
- |
- |
CDK, putative [Ricinus communis] |
| 11 |
Hb_007044_260 |
0.109927425 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 12 |
Hb_011063_030 |
0.1120258412 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_008132_010 |
0.1120700409 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 14 |
Hb_119044_040 |
0.1142176949 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 15 |
Hb_001541_140 |
0.1150198226 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 16 |
Hb_001279_110 |
0.1167960673 |
- |
- |
PREDICTED: uncharacterized protein LOC105633338 isoform X4 [Jatropha curcas] |
| 17 |
Hb_002928_150 |
0.1199674716 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |
| 18 |
Hb_003398_050 |
0.1201546763 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 19 |
Hb_003776_050 |
0.121778363 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 20 |
Hb_001998_180 |
0.1226230151 |
- |
- |
actin binding protein, putative [Ricinus communis] |