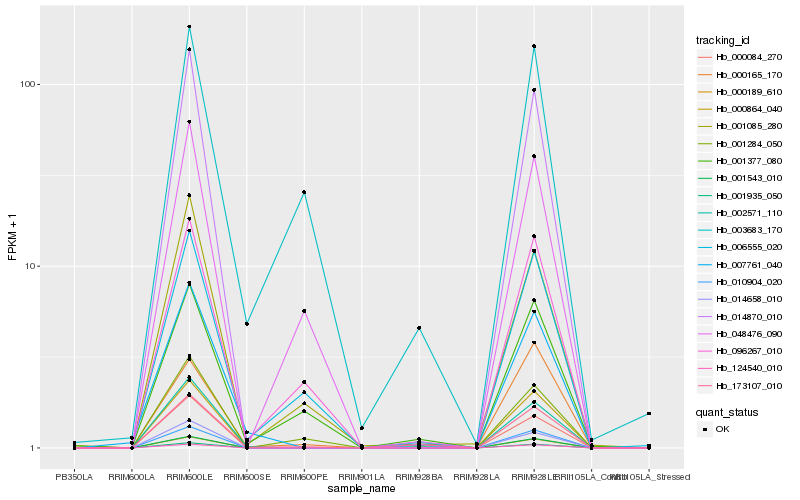
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_173107_010 |
0.0 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 2 |
Hb_002571_110 |
0.1072286614 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 3 |
Hb_001377_080 |
0.1219334269 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 4 |
Hb_006555_020 |
0.1226025022 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 5 |
Hb_001284_050 |
0.1303837917 |
- |
- |
PREDICTED: uncharacterized protein LOC100781779 isoform X1 [Glycine max] |
| 6 |
Hb_014658_010 |
0.1315241598 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 7 |
Hb_007761_040 |
0.132574718 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 8 |
Hb_001935_050 |
0.1365645542 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 [Jatropha curcas] |
| 9 |
Hb_000864_040 |
0.1380280269 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000189_610 |
0.1387438564 |
- |
- |
Late embryogenesis abundant protein D-34, putative [Ricinus communis] |
| 11 |
Hb_001543_010 |
0.1389600914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010904_020 |
0.140209457 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 [Populus euphratica] |
| 13 |
Hb_124540_010 |
0.140755551 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1-like [Populus euphratica] |
| 14 |
Hb_014870_010 |
0.140768794 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_001085_280 |
0.1417901786 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 16 |
Hb_000165_170 |
0.1428722054 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 17 |
Hb_096267_010 |
0.144173031 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 18 |
Hb_003683_170 |
0.1474505579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000084_270 |
0.1478039209 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 20 |
Hb_048476_090 |
0.1482974604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |