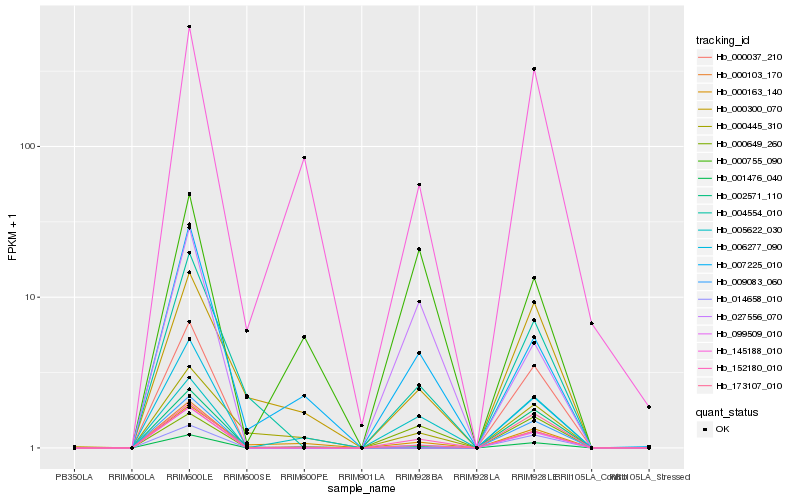
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_152180_010 |
0.0 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 2 |
Hb_014658_010 |
0.1075468938 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 3 |
Hb_004554_010 |
0.1671899994 |
- |
- |
PREDICTED: uncharacterized protein LOC105636233 isoform X1 [Jatropha curcas] |
| 4 |
Hb_027556_070 |
0.169129802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001476_040 |
0.1895588487 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_007225_010 |
0.1904332578 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 7 |
Hb_005622_030 |
0.1971799438 |
- |
- |
transferase, putative [Ricinus communis] |
| 8 |
Hb_000163_140 |
0.2149716261 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 9 |
Hb_173107_010 |
0.2225733178 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 10 |
Hb_000649_260 |
0.2257742562 |
- |
- |
uncharacterized LOC105634108 [Jatropha curcas] |
| 11 |
Hb_000755_090 |
0.2285961109 |
- |
- |
transferase, putative [Ricinus communis] |
| 12 |
Hb_002571_110 |
0.2358507466 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 13 |
Hb_000300_070 |
0.2362736266 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 14 |
Hb_099509_010 |
0.2401796655 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000445_310 |
0.241090587 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 16 |
Hb_000103_170 |
0.2416561163 |
- |
- |
uncharacterized protein LOC100501254 [Zea mays] |
| 17 |
Hb_145188_010 |
0.2422537863 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 18 |
Hb_006277_090 |
0.2426146976 |
- |
- |
PREDICTED: uncharacterized protein LOC105649700 [Jatropha curcas] |
| 19 |
Hb_009083_060 |
0.2440726001 |
- |
- |
PREDICTED: uncharacterized protein LOC103344557 [Prunus mume] |
| 20 |
Hb_000037_210 |
0.2442254375 |
- |
- |
- |