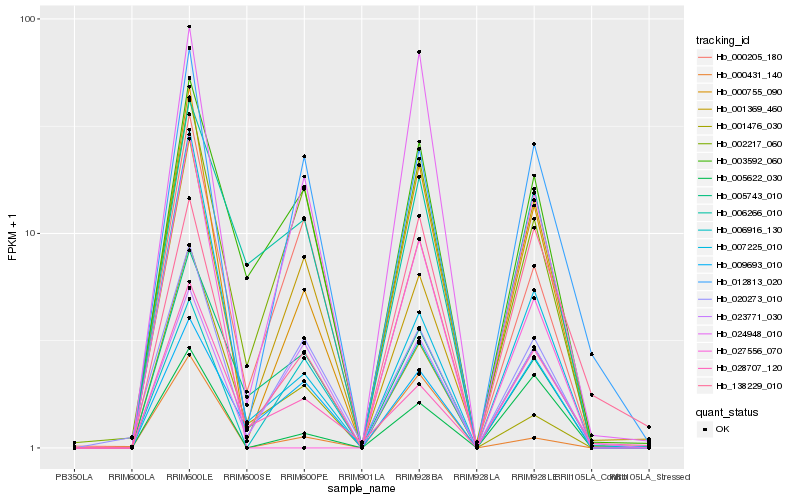
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000755_090 |
0.0 |
- |
- |
transferase, putative [Ricinus communis] |
| 2 |
Hb_024948_010 |
0.1600127044 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 3 |
Hb_005622_030 |
0.1652365978 |
- |
- |
transferase, putative [Ricinus communis] |
| 4 |
Hb_020273_010 |
0.1697086402 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 5 |
Hb_012813_020 |
0.1748478775 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_002217_060 |
0.1871542681 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 7 |
Hb_027556_070 |
0.1899995664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006916_130 |
0.1903680049 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 9 |
Hb_003592_060 |
0.1926501599 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |
| 10 |
Hb_005743_010 |
0.1967916181 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 11 |
Hb_000431_140 |
0.2012001737 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 12 |
Hb_028707_120 |
0.2042286392 |
- |
- |
PREDICTED: uncharacterized protein LOC105645799 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000205_180 |
0.2083644748 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 14 |
Hb_007225_010 |
0.2087859402 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 15 |
Hb_138229_010 |
0.2097103579 |
- |
- |
hypothetical protein JCGZ_05736 [Jatropha curcas] |
| 16 |
Hb_001476_030 |
0.2118268544 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 17 |
Hb_023771_030 |
0.2118930656 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_009693_010 |
0.2123731777 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |
| 19 |
Hb_001369_460 |
0.2136964721 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 20 |
Hb_006266_010 |
0.2153836933 |
- |
- |
unnamed protein product [Vitis vinifera] |