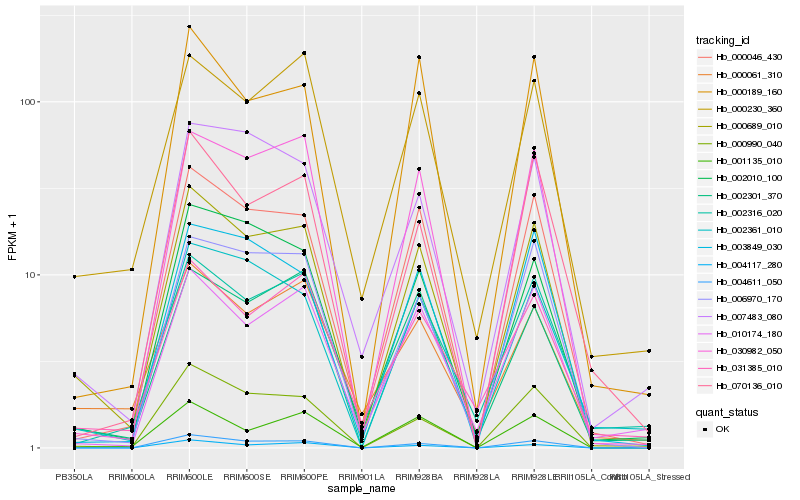
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000689_010 |
0.0 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 2 |
Hb_000046_430 |
0.0966728942 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 3 |
Hb_000990_040 |
0.1084893538 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 4 |
Hb_004611_050 |
0.1138720865 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 5 |
Hb_006970_170 |
0.1147403151 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_030982_050 |
0.1150687727 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 7 |
Hb_002010_100 |
0.1180635931 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 8 |
Hb_001135_010 |
0.125055265 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 9 |
Hb_002361_010 |
0.125865706 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_002301_370 |
0.1260087457 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 11 |
Hb_000189_160 |
0.1260578779 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 12 |
Hb_000230_360 |
0.1275187229 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 13 |
Hb_007483_080 |
0.1283965763 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 14 |
Hb_002316_020 |
0.1310530416 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 15 |
Hb_003849_030 |
0.1313269312 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 16 |
Hb_010174_180 |
0.1327424718 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_000061_310 |
0.1336210364 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 18 |
Hb_070136_010 |
0.1338736241 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 19 |
Hb_031385_010 |
0.1360748816 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 20 |
Hb_004117_280 |
0.136139728 |
- |
- |
hypothetical protein JCGZ_00361 [Jatropha curcas] |