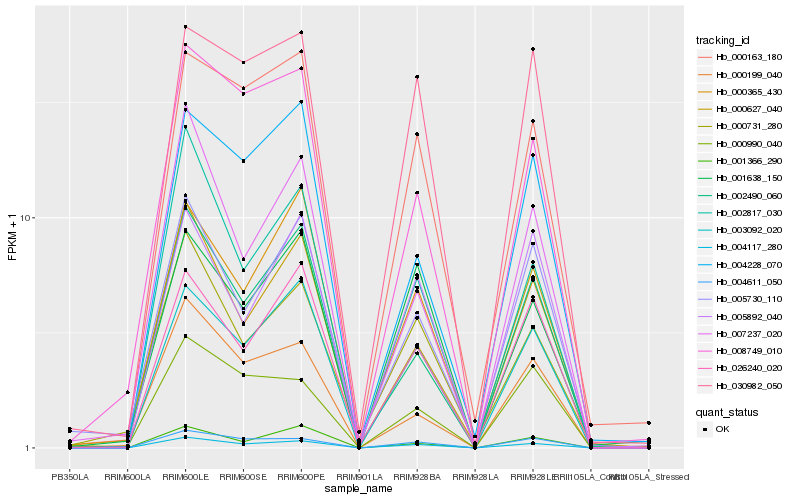
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004117_280 |
0.0 |
- |
- |
hypothetical protein JCGZ_00361 [Jatropha curcas] |
| 2 |
Hb_004611_050 |
0.0804344071 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 3 |
Hb_003092_020 |
0.0867742808 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 4 |
Hb_002817_030 |
0.1017151463 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 5 |
Hb_002490_060 |
0.1061680543 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 6 |
Hb_000627_040 |
0.1105231872 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 7 |
Hb_000163_180 |
0.1127774839 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 8 |
Hb_008749_010 |
0.1161009834 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 9 |
Hb_026240_020 |
0.1174906693 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 10 |
Hb_000731_280 |
0.1196681455 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000365_430 |
0.1216034126 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 12 |
Hb_000199_040 |
0.1220661234 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_030982_050 |
0.124322784 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 14 |
Hb_005730_110 |
0.1264710183 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 15 |
Hb_007237_020 |
0.1275162006 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 16 |
Hb_001366_290 |
0.1288492538 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000990_040 |
0.1291692089 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 18 |
Hb_001638_150 |
0.1311310809 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 19 |
Hb_005892_040 |
0.1311725332 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 20 |
Hb_004228_070 |
0.1322359904 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |