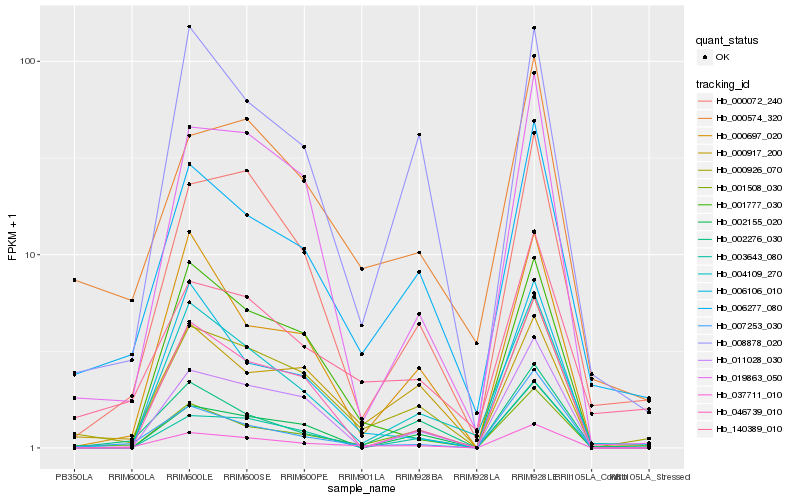
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_037711_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_000926_070 |
0.1385182493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007253_030 |
0.1460319382 |
- |
- |
PREDICTED: 33 kDa ribonucleoprotein, chloroplastic-like [Populus euphratica] |
| 4 |
Hb_006277_080 |
0.1507396159 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 5 |
Hb_001508_030 |
0.1550700823 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |
| 6 |
Hb_019863_050 |
0.1609922589 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 7 |
Hb_000072_240 |
0.1624933832 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000697_020 |
0.1667144696 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_001777_030 |
0.1670377815 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 10 |
Hb_002276_030 |
0.1690529339 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 11 |
Hb_046739_010 |
0.1709805243 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 12 |
Hb_011028_030 |
0.1730571551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002155_020 |
0.1750715197 |
- |
- |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 14 |
Hb_008878_020 |
0.1763963195 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 15 |
Hb_003643_080 |
0.1767377582 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 16 |
Hb_004109_270 |
0.1772490464 |
transcription factor |
TF Family: NAC |
transcription activator NAC1 family protein [Populus trichocarpa] |
| 17 |
Hb_000574_320 |
0.1782644835 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 18 |
Hb_000917_200 |
0.1784006248 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 19 |
Hb_140389_010 |
0.179324332 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 20 |
Hb_006106_010 |
0.1793568135 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |