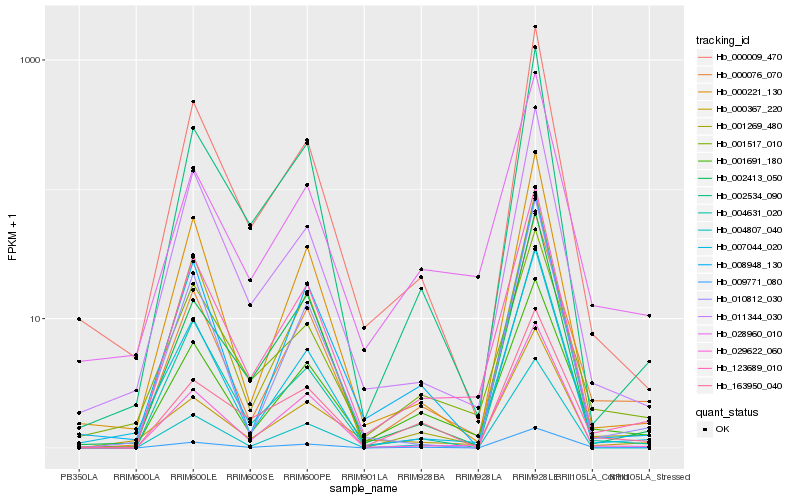
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000076_070 |
0.0 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 2 |
Hb_009771_080 |
0.0674403633 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 3 |
Hb_001691_180 |
0.0787291718 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 4 |
Hb_123689_010 |
0.0989141063 |
- |
- |
PREDICTED: uncharacterized protein At1g18480 [Jatropha curcas] |
| 5 |
Hb_029622_060 |
0.1032014538 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007044_020 |
0.1088440294 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000221_130 |
0.1118382526 |
- |
- |
thylakoid lumenal 29 kDa protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008948_130 |
0.1173440894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010812_030 |
0.1174204323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001269_480 |
0.1175985329 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_028960_010 |
0.1189830791 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 12 |
Hb_000009_470 |
0.1244578656 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_002413_050 |
0.125330816 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 14 |
Hb_002534_090 |
0.1263814518 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 15 |
Hb_000367_220 |
0.1265055923 |
- |
- |
hypothetical protein RCOM_1470580 [Ricinus communis] |
| 16 |
Hb_004631_020 |
0.1280520227 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 17 |
Hb_011344_030 |
0.1305519049 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004807_040 |
0.1310206057 |
- |
- |
PREDICTED: uncharacterized protein LOC104420559 [Eucalyptus grandis] |
| 19 |
Hb_163950_040 |
0.1313160666 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001517_010 |
0.1314928576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |