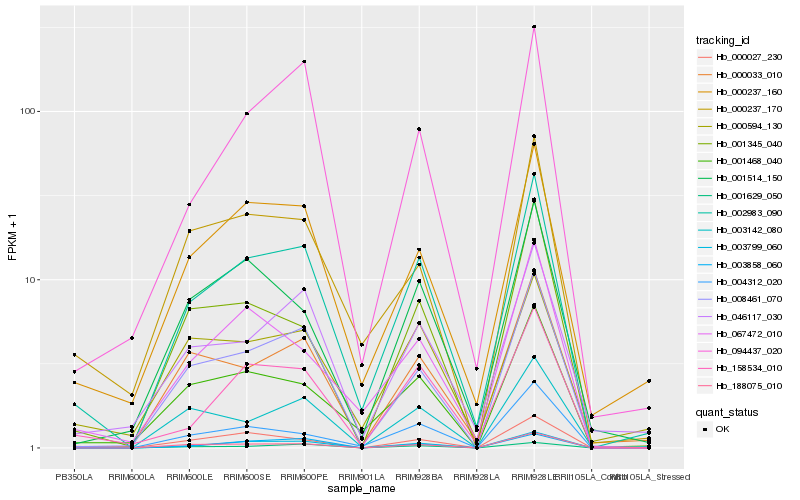
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002983_090 |
0.0 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |
| 2 |
Hb_000237_160 |
0.1161110678 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |
| 3 |
Hb_001468_040 |
0.124329689 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 4 |
Hb_008461_070 |
0.1257798895 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_158534_010 |
0.1285825765 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_046117_030 |
0.1313837722 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000033_010 |
0.1316482339 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 8 |
Hb_067472_010 |
0.1346299048 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 9 |
Hb_188075_010 |
0.1384198565 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 10 |
Hb_000594_130 |
0.1423674709 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_094437_020 |
0.1424328329 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_003142_080 |
0.1435056122 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 13 |
Hb_000027_230 |
0.1452815233 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_000237_170 |
0.1487921304 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 15 |
Hb_004312_020 |
0.1489553405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001514_150 |
0.1530542016 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 17 |
Hb_001345_040 |
0.1554488616 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 18 |
Hb_003858_060 |
0.1586956135 |
- |
- |
Germin-like protein subfamily 1 member 18 [Theobroma cacao] |
| 19 |
Hb_001629_050 |
0.1587434732 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 20 |
Hb_003799_060 |
0.1598865753 |
- |
- |
hypothetical protein POPTR_0005s22800g [Populus trichocarpa] |