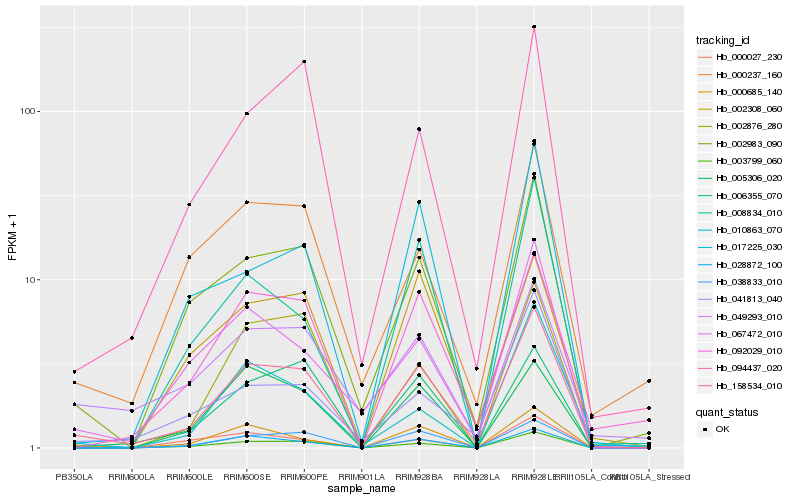
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158534_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_002983_090 |
0.1285825765 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |
| 3 |
Hb_038833_010 |
0.1458086568 |
- |
- |
polyprotein [Ananas comosus] |
| 4 |
Hb_028872_100 |
0.1533669145 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 5 |
Hb_094437_020 |
0.1548822964 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_092029_010 |
0.1579196356 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 7 |
Hb_067472_010 |
0.1667441275 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 8 |
Hb_000237_160 |
0.1703059295 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |
| 9 |
Hb_000685_140 |
0.172306586 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 10 |
Hb_041813_040 |
0.1725600032 |
- |
- |
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain 5, truncated - sugar beet mitochondrion (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_005306_020 |
0.1754196195 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 12 |
Hb_049293_010 |
0.1756387072 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_017225_030 |
0.175726121 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 14 |
Hb_006355_070 |
0.176671533 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 15 |
Hb_002876_280 |
0.1770218093 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 16 |
Hb_002308_060 |
0.1775451563 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 17 |
Hb_000027_230 |
0.1778585823 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_003799_060 |
0.1820328315 |
- |
- |
hypothetical protein POPTR_0005s22800g [Populus trichocarpa] |
| 19 |
Hb_010863_070 |
0.1822792101 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 20 |
Hb_008834_010 |
0.1825021325 |
- |
- |
hypothetical protein POPTR_0016s04660g [Populus trichocarpa] |