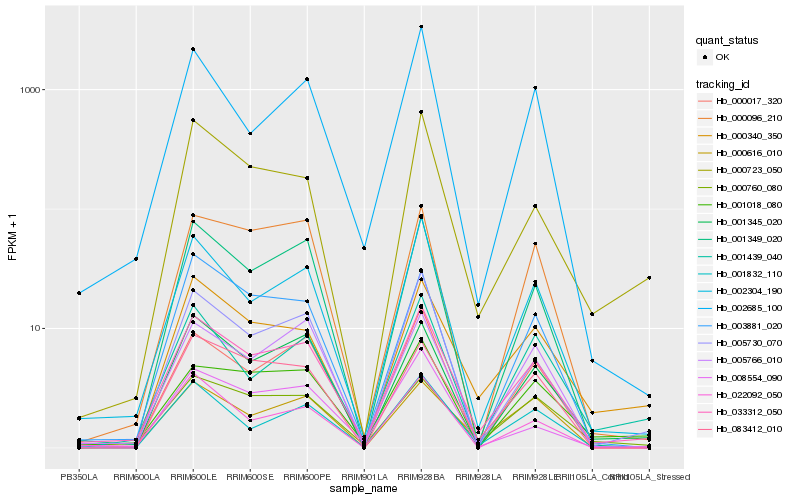
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033312_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 2 |
Hb_002304_190 |
0.0870583976 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |
| 3 |
Hb_000760_080 |
0.1115826978 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 4 |
Hb_001349_020 |
0.1126673986 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 5 |
Hb_001832_110 |
0.1133629207 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005730_070 |
0.1173313063 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 7 |
Hb_000017_320 |
0.1195446186 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor GLABRA 3-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000723_050 |
0.12355286 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 9 |
Hb_001345_020 |
0.1247962836 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 10 |
Hb_002685_100 |
0.1270674247 |
- |
- |
low temperature inducible SLTI66 [Glycine max] |
| 11 |
Hb_000340_350 |
0.127109791 |
- |
- |
PREDICTED: sugar transporter ERD6-like 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_083412_010 |
0.1292368321 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 13 |
Hb_005766_010 |
0.1302684077 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_001018_080 |
0.1320046624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003881_020 |
0.1324404321 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 16 |
Hb_022092_050 |
0.1337937298 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 17 |
Hb_000616_010 |
0.1347203867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001439_040 |
0.1350104085 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_008554_090 |
0.1361579599 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000096_210 |
0.1370095854 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |