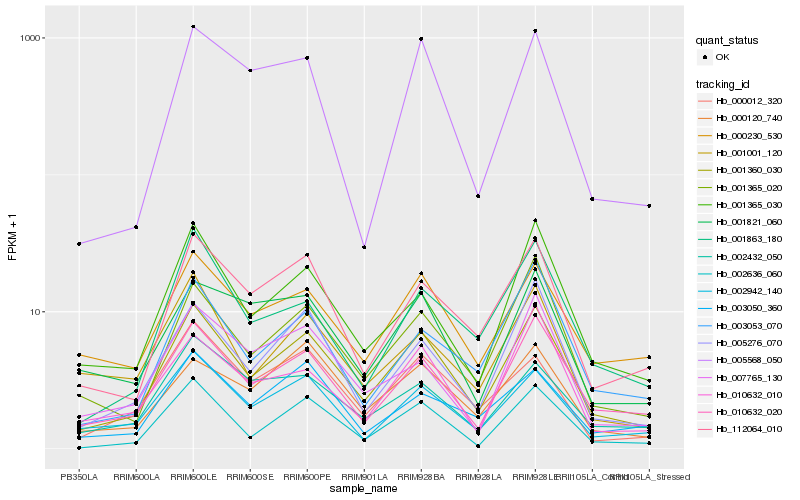
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001360_030 |
0.0 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 2 |
Hb_010632_020 |
0.0923295264 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 3 |
Hb_001365_020 |
0.1108914036 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 4 |
Hb_002636_060 |
0.111810979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_112064_010 |
0.1119201423 |
- |
- |
PREDICTED: inositol-phosphate phosphatase [Jatropha curcas] |
| 6 |
Hb_005276_070 |
0.1141747089 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 7 |
Hb_001365_030 |
0.1147752441 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 8 |
Hb_002942_140 |
0.1248395897 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 9 |
Hb_003053_070 |
0.1288678463 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000230_530 |
0.129095681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001863_180 |
0.1342476424 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000120_740 |
0.137478201 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007765_130 |
0.1382099319 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000012_320 |
0.1390460174 |
- |
- |
PREDICTED: uncharacterized protein LOC105638164 [Jatropha curcas] |
| 15 |
Hb_001001_120 |
0.1397937523 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 16 |
Hb_005568_050 |
0.140338474 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 17 |
Hb_002432_050 |
0.1428716031 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 18 |
Hb_010632_010 |
0.1436436416 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_003050_360 |
0.1443325683 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001821_060 |
0.1444502242 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |