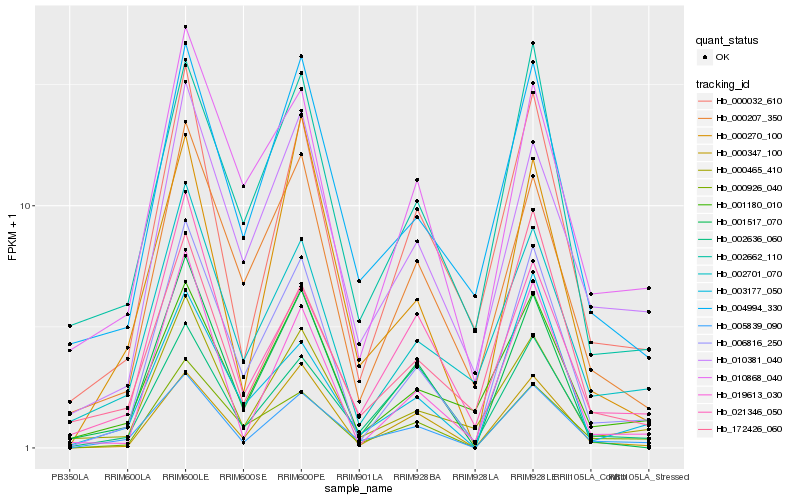
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005839_090 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g56230 isoform X1 [Populus euphratica] |
| 2 |
Hb_006816_250 |
0.1329118388 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 3 |
Hb_000032_610 |
0.1331441639 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000926_040 |
0.1347374729 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH5 [Jatropha curcas] |
| 5 |
Hb_000270_100 |
0.1397694556 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 6 |
Hb_002636_060 |
0.1431386517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000347_100 |
0.1521439855 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |
| 8 |
Hb_001517_070 |
0.1521952315 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_003177_050 |
0.1563274123 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Populus euphratica] |
| 10 |
Hb_021346_050 |
0.1585784575 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 11 |
Hb_000465_410 |
0.1607162343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004994_330 |
0.160923396 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_010381_040 |
0.1613158643 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 14 |
Hb_019613_030 |
0.1647026616 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 15 |
Hb_001180_010 |
0.166268063 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 16 |
Hb_002662_110 |
0.166701909 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 17 |
Hb_002701_070 |
0.1679112846 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 18 |
Hb_172426_060 |
0.1681661344 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 19 |
Hb_010868_040 |
0.1698914766 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 20 |
Hb_000207_350 |
0.1706092044 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |