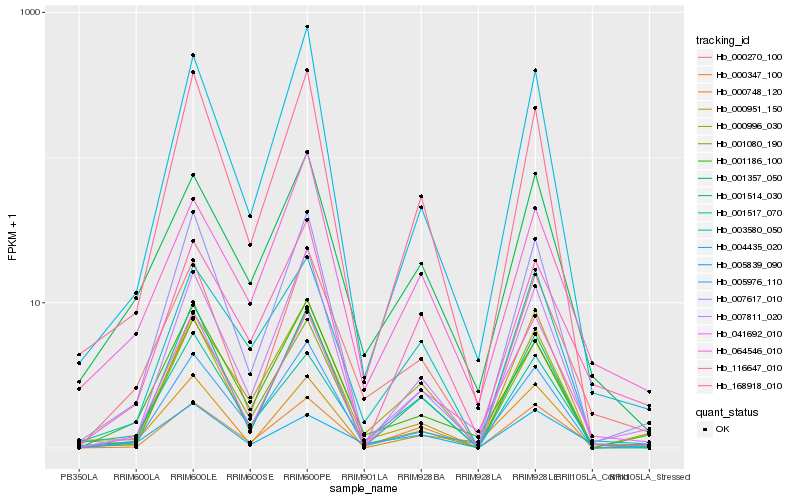
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_100 |
0.0 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 2 |
Hb_001357_050 |
0.1265043328 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |
| 3 |
Hb_000347_100 |
0.1382587932 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |
| 4 |
Hb_005839_090 |
0.1397694556 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g56230 isoform X1 [Populus euphratica] |
| 5 |
Hb_041692_010 |
0.1451198241 |
- |
- |
annexin [Manihot esculenta] |
| 6 |
Hb_007811_020 |
0.1474960136 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 7 |
Hb_001186_100 |
0.1498499718 |
- |
- |
PREDICTED: uncharacterized protein LOC105640806 isoform X1 [Jatropha curcas] |
| 8 |
Hb_168918_010 |
0.1519379377 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 9 |
Hb_064546_010 |
0.1545779476 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 10 |
Hb_005976_110 |
0.157549281 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 11 |
Hb_001514_030 |
0.1592615392 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 12 |
Hb_004435_020 |
0.1593949348 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 13 |
Hb_007617_010 |
0.1596410028 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003580_050 |
0.1610745524 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 15 |
Hb_116647_010 |
0.1617826281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001080_190 |
0.1624808494 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001517_070 |
0.1632020648 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000996_030 |
0.1633374513 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000748_120 |
0.1638241851 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 20 |
Hb_000951_150 |
0.1644717176 |
- |
- |
hypothetical protein POPTR_0015s05850g [Populus trichocarpa] |