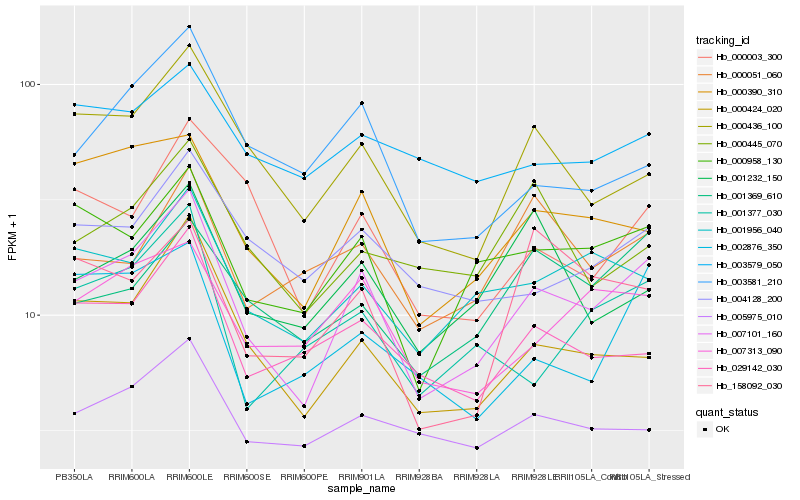
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007101_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000424_020 |
0.1193832601 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |
| 3 |
Hb_000390_310 |
0.1205589769 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000958_130 |
0.1215523501 |
- |
- |
PREDICTED: uncharacterized protein LOC105628716 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000436_100 |
0.1293314702 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 6 |
Hb_001956_040 |
0.1333219449 |
- |
- |
PREDICTED: PGR5-like protein 1B, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_007313_090 |
0.1340768568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_029142_030 |
0.1386732028 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 9 |
Hb_005975_010 |
0.1428405675 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_004128_200 |
0.1444165728 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 11 |
Hb_000003_300 |
0.1448189208 |
- |
- |
PREDICTED: uncharacterized protein LOC105631182 [Jatropha curcas] |
| 12 |
Hb_001377_030 |
0.1451170371 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 13 |
Hb_001369_610 |
0.148151715 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003581_210 |
0.1485020779 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-binding protein WHY1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002876_350 |
0.1504390902 |
- |
- |
PREDICTED: gibberellin-regulated protein 9-like isoform X2 [Citrus sinensis] |
| 16 |
Hb_001232_150 |
0.1517645029 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 17 |
Hb_000445_070 |
0.1561381587 |
- |
- |
PREDICTED: uncharacterized protein LOC105638998 [Jatropha curcas] |
| 18 |
Hb_000051_060 |
0.157592837 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_158092_030 |
0.1588468069 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP19, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003579_050 |
0.1604970805 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |