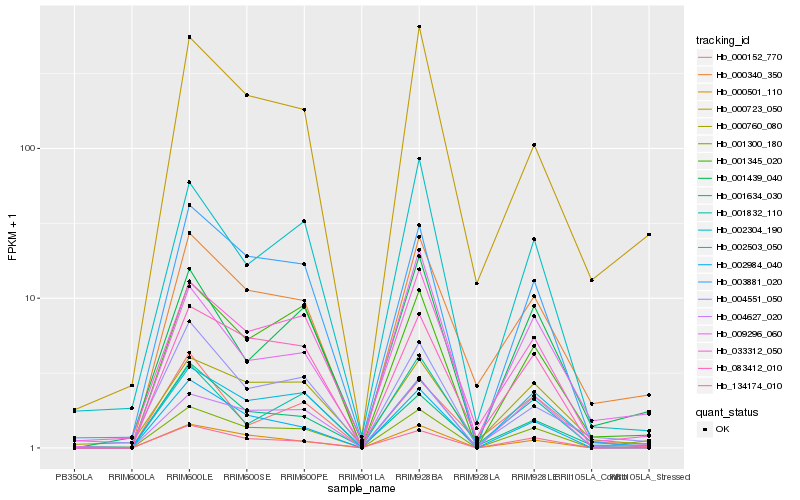
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_350 |
0.0 |
- |
- |
PREDICTED: sugar transporter ERD6-like 7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000723_050 |
0.1085104062 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 3 |
Hb_033312_050 |
0.127109791 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 4 |
Hb_000760_080 |
0.1434229211 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 5 |
Hb_001634_030 |
0.1450770405 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |
| 6 |
Hb_001300_180 |
0.1453206481 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 7 |
Hb_134174_010 |
0.146802274 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_083412_010 |
0.1482641576 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 9 |
Hb_000152_770 |
0.1524389986 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 10 |
Hb_002503_050 |
0.1567443098 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 11 |
Hb_004551_050 |
0.1579102408 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001345_020 |
0.1620455491 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 13 |
Hb_001439_040 |
0.1646604225 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003881_020 |
0.1651621526 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 15 |
Hb_002984_040 |
0.1651631043 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000501_110 |
0.1660126196 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 17 |
Hb_004627_020 |
0.1718543613 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 18 |
Hb_009296_060 |
0.1727885608 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 19 |
Hb_001832_110 |
0.1741036534 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002304_190 |
0.1741852841 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |