| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
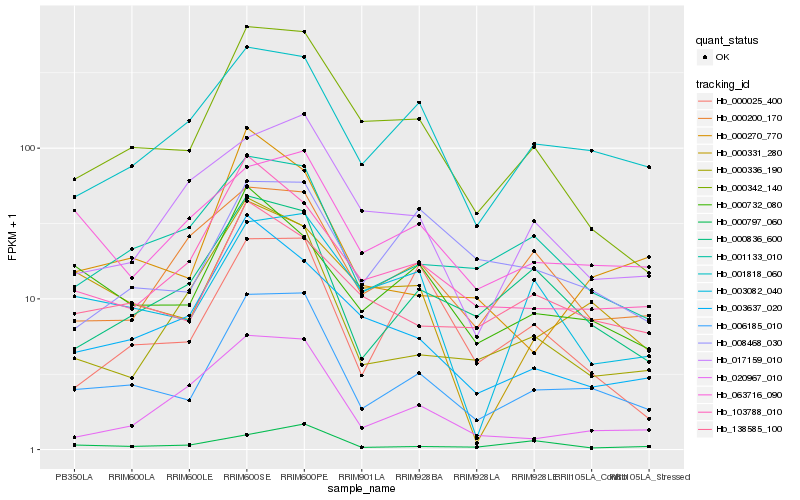
Hb_006185_010 |
0.0 |
- |
- |
hypothetical protein L484_013453 [Morus notabilis] |
| 2 |
Hb_000342_140 |
0.1507900319 |
- |
- |
PREDICTED: vacuolar-processing enzyme-like [Jatropha curcas] |
| 3 |
Hb_001818_060 |
0.1539857458 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 4 |
Hb_000836_600 |
0.1560512463 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 5 |
Hb_001133_010 |
0.1611054537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_138585_100 |
0.1733032253 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 7 |
Hb_103788_010 |
0.1737185896 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_000732_080 |
0.1817548266 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |
| 9 |
Hb_000331_280 |
0.1822656977 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 10 |
Hb_063716_090 |
0.1829861374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000797_060 |
0.1845307132 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 12 |
Hb_000270_770 |
0.1849002427 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 13 |
Hb_003082_040 |
0.1857739002 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Phoenix dactylifera] |
| 14 |
Hb_000025_400 |
0.1893947077 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 15 |
Hb_000200_170 |
0.1925933821 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 16 |
Hb_020967_010 |
0.1932659586 |
- |
- |
- |
| 17 |
Hb_017159_010 |
0.1944729521 |
- |
- |
- |
| 18 |
Hb_008468_030 |
0.1952991998 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 19 |
Hb_003637_020 |
0.1958239938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000336_190 |
0.1968474045 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |