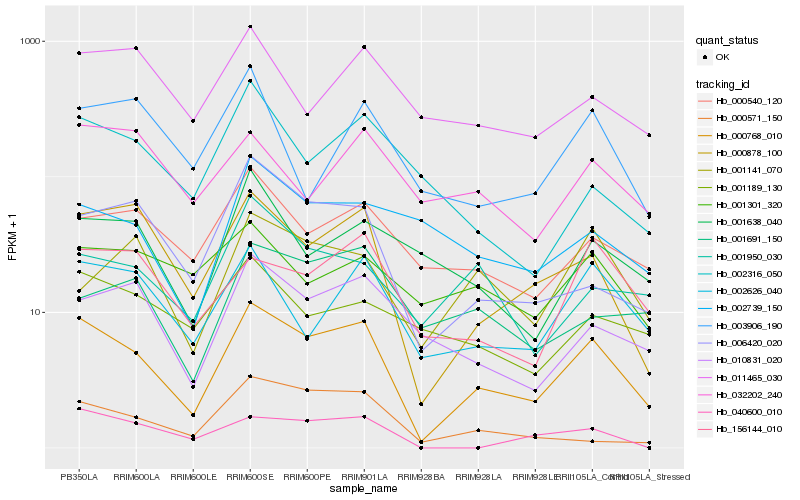
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000768_010 |
0.0 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X3 [Citrus sinensis] |
| 2 |
Hb_000878_100 |
0.1641914824 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 3 |
Hb_002626_040 |
0.1873264632 |
- |
- |
PREDICTED: inter-alpha-trypsin inhibitor heavy chain H3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006420_020 |
0.1994851773 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 5 |
Hb_156144_010 |
0.2028832009 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 6 |
Hb_000540_120 |
0.2030970942 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 7 |
Hb_010831_020 |
0.2041601629 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_000571_150 |
0.2176735586 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001950_030 |
0.2188566845 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 10 |
Hb_011465_030 |
0.2199579418 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 11 |
Hb_001638_040 |
0.2206132771 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 12 |
Hb_002316_050 |
0.2206338363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_040600_010 |
0.2212525602 |
- |
- |
hypothetical protein VITISV_036432 [Vitis vinifera] |
| 14 |
Hb_001141_070 |
0.2215936692 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 3 [Jatropha curcas] |
| 15 |
Hb_002739_150 |
0.2242162126 |
- |
- |
Uncharacterized protein TCM_016791 [Theobroma cacao] |
| 16 |
Hb_001691_150 |
0.2256284126 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Jatropha curcas] |
| 17 |
Hb_001189_130 |
0.2256734572 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 18 |
Hb_032202_240 |
0.2264281586 |
- |
- |
PREDICTED: F-box protein At4g00755 [Jatropha curcas] |
| 19 |
Hb_003906_190 |
0.2264644772 |
- |
- |
hypothetical protein EUGRSUZ_K00213 [Eucalyptus grandis] |
| 20 |
Hb_001301_320 |
0.227496957 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |