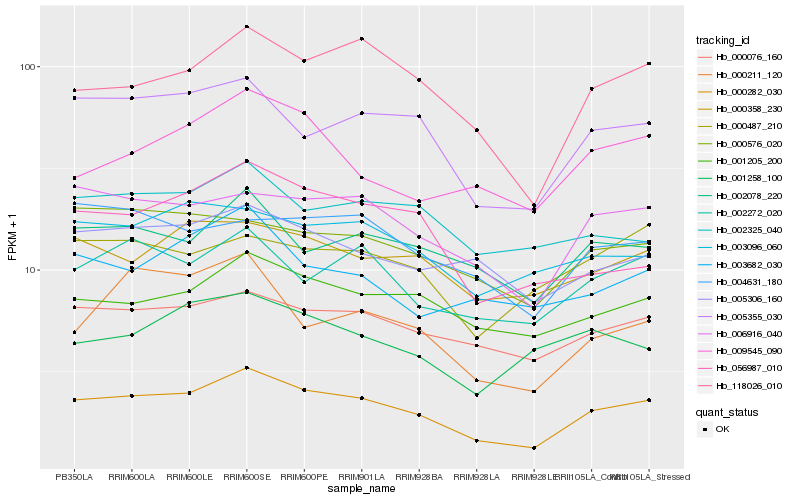
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000282_030 |
0.0 |
- |
- |
K(+) efflux antiporter 4 [Glycine soja] |
| 2 |
Hb_118026_010 |
0.0829768163 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 3 |
Hb_005355_030 |
0.0845064404 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 4 |
Hb_006916_040 |
0.0845281788 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 5 |
Hb_000076_160 |
0.0934255201 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 6 |
Hb_001205_200 |
0.0966518073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002272_020 |
0.0979941724 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 8 |
Hb_002078_220 |
0.0997214773 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000358_230 |
0.1017189577 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 10 |
Hb_005306_160 |
0.1031157583 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_004631_180 |
0.1047279484 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 12 |
Hb_000576_020 |
0.1051802386 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 13 |
Hb_003096_060 |
0.1052797518 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 14 |
Hb_056987_010 |
0.1092685885 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 15 |
Hb_002325_040 |
0.1105274651 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009545_090 |
0.1123777752 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 17 |
Hb_003682_030 |
0.1126177831 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 18 |
Hb_000487_210 |
0.1136315284 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 19 |
Hb_000211_120 |
0.114424353 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 20 |
Hb_001258_100 |
0.1151629613 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |