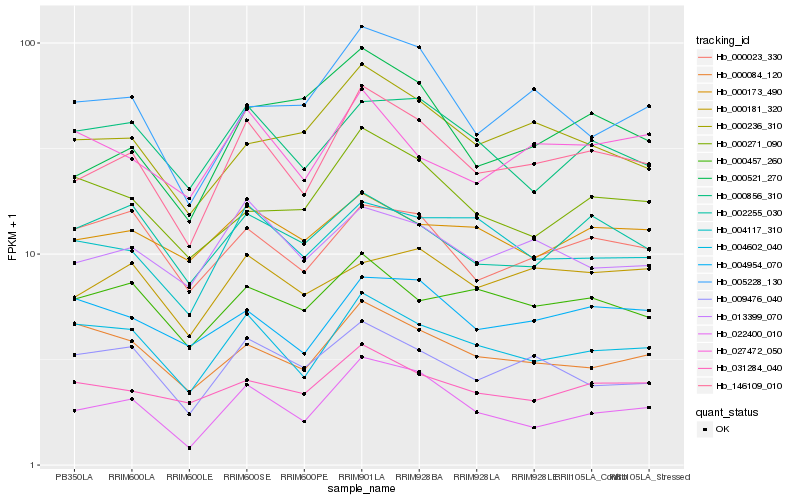
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146109_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_022400_010 |
0.084077498 |
- |
- |
Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase family protein [Theobroma cacao] |
| 3 |
Hb_004602_040 |
0.0864353911 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_013399_070 |
0.0959489953 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 5 |
Hb_031284_040 |
0.1026142247 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g30700 [Jatropha curcas] |
| 6 |
Hb_004117_310 |
0.1033119547 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 7 |
Hb_000236_310 |
0.103630174 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Malus domestica] |
| 8 |
Hb_009476_040 |
0.1046458708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000457_260 |
0.1065918232 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_005228_130 |
0.1085268597 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_000023_330 |
0.1090536481 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 12 |
Hb_004954_070 |
0.1095123987 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 13 |
Hb_000856_310 |
0.1097018305 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 14 |
Hb_027472_050 |
0.1108626401 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 15 |
Hb_000181_320 |
0.1109475366 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 16 |
Hb_000521_270 |
0.1116465265 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 17 |
Hb_000173_490 |
0.1127731858 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002255_030 |
0.1132055587 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 19 |
Hb_000084_120 |
0.1132963586 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g36240 [Jatropha curcas] |
| 20 |
Hb_000271_090 |
0.1147482342 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |