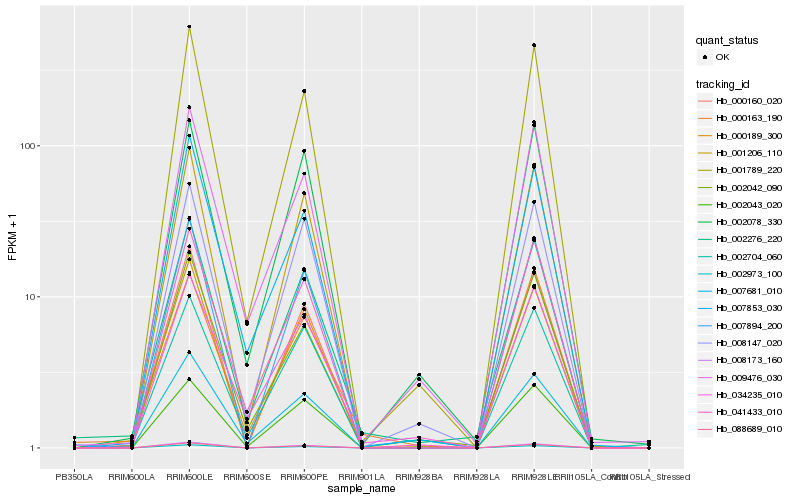
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002276_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000189_300 |
0.0791087351 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_001789_220 |
0.0836082375 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 4 |
Hb_034235_010 |
0.0841195733 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_007681_010 |
0.0863493202 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 6 |
Hb_041433_010 |
0.0878529345 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_001206_110 |
0.0894611632 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 8 |
Hb_002704_060 |
0.0905212505 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 9 |
Hb_000163_190 |
0.0905311682 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 10 |
Hb_008147_020 |
0.090946439 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 11 |
Hb_002042_090 |
0.0920098977 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 12 |
Hb_009476_030 |
0.0928471729 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 13 |
Hb_002043_020 |
0.0941186528 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 14 |
Hb_007894_200 |
0.09515216 |
- |
- |
ubiquitin-conjugating enzyme variant, putative [Ricinus communis] |
| 15 |
Hb_007853_030 |
0.0962681519 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 16 |
Hb_000160_020 |
0.0962857117 |
- |
- |
PREDICTED: gamma-interferon-inducible lysosomal thiol reductase-like [Jatropha curcas] |
| 17 |
Hb_002078_330 |
0.0971023027 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 18 |
Hb_088689_010 |
0.0995014249 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 19 |
Hb_008173_160 |
0.0995080707 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 20 |
Hb_002973_100 |
0.0998836773 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable N-acetyltransferase HLS1 [Populus euphratica] |