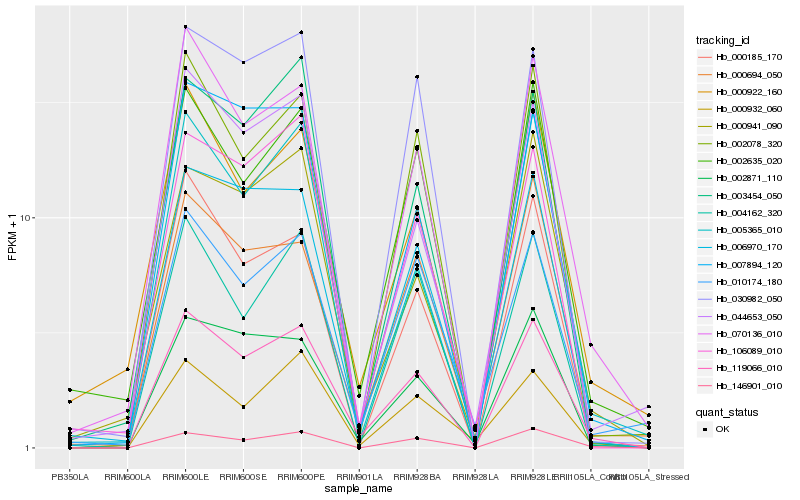
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_119066_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 2 |
Hb_070136_010 |
0.0955229102 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 3 |
Hb_002871_110 |
0.104098072 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 4 |
Hb_006970_170 |
0.1114531127 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_002078_320 |
0.1118255865 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_002635_020 |
0.112200713 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 7 |
Hb_146901_010 |
0.1128832548 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 8 |
Hb_000185_170 |
0.1174314664 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_007894_120 |
0.1191956009 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_044653_050 |
0.1208031924 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005365_010 |
0.1212837363 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 12 |
Hb_000694_050 |
0.1213468652 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 13 |
Hb_030982_050 |
0.1223973456 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 14 |
Hb_003454_050 |
0.1234080417 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 15 |
Hb_004162_320 |
0.1244669601 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000922_160 |
0.1250473649 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 17 |
Hb_106089_010 |
0.1254934871 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 18 |
Hb_000932_060 |
0.1257457411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010174_180 |
0.1259960496 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_000941_090 |
0.1281245505 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |