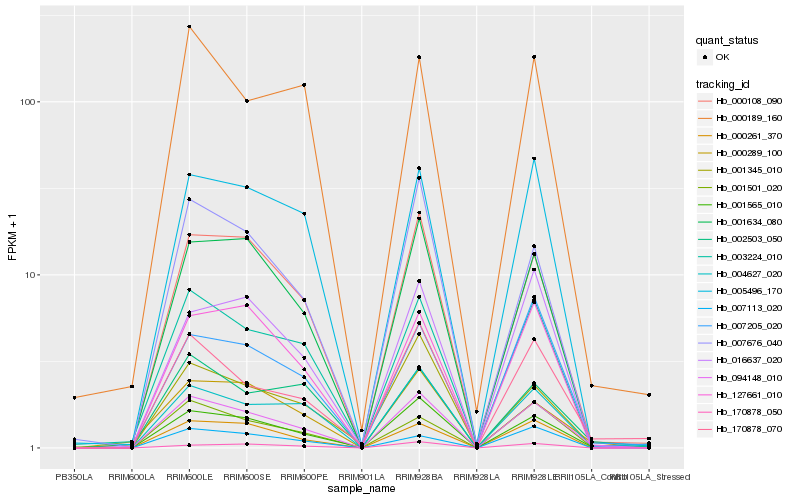
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094148_010 |
0.0 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001565_010 |
0.0943871405 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 3 |
Hb_000108_090 |
0.1042789302 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 4 |
Hb_003224_010 |
0.1063979334 |
- |
- |
- |
| 5 |
Hb_001634_080 |
0.1084044759 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000261_370 |
0.1131304667 |
- |
- |
- |
| 7 |
Hb_170878_070 |
0.1156165034 |
- |
- |
hypothetical protein EUGRSUZ_L008133, partial [Eucalyptus grandis] |
| 8 |
Hb_004627_020 |
0.1228966125 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 9 |
Hb_005496_170 |
0.123561683 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 10 |
Hb_007676_040 |
0.1238719112 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 11 |
Hb_127661_010 |
0.133245672 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001345_010 |
0.1338510346 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |
| 13 |
Hb_001501_020 |
0.1340480038 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 14 |
Hb_000289_100 |
0.1359388844 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 15 |
Hb_007205_020 |
0.140131471 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 16 |
Hb_002503_050 |
0.1462179701 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 17 |
Hb_170878_050 |
0.146603689 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 18 |
Hb_000189_160 |
0.148147411 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 19 |
Hb_016637_020 |
0.1484567047 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 20 |
Hb_007113_020 |
0.1535918765 |
- |
- |
- |