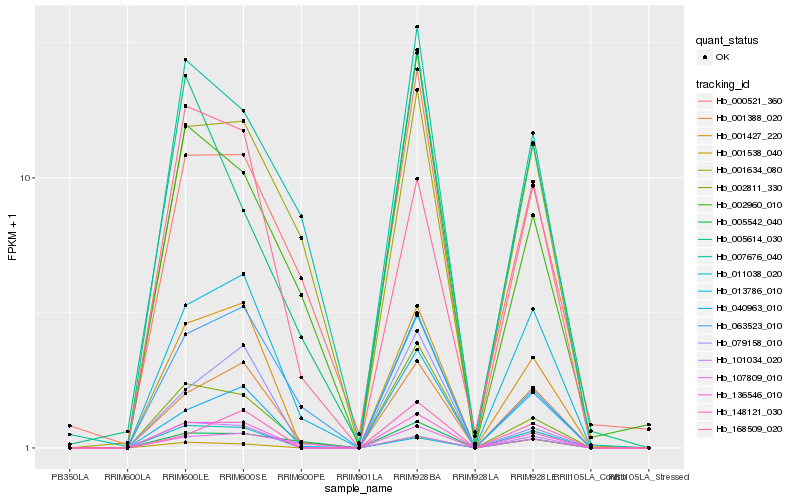
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_136546_010 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1, partial [Populus euphratica] |
| 2 |
Hb_001427_220 |
0.0614864152 |
- |
- |
hypothetical protein CISIN_1g033375mg [Citrus sinensis] |
| 3 |
Hb_001388_020 |
0.135608496 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP20 [Jatropha curcas] |
| 4 |
Hb_002811_330 |
0.1399717174 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 5 |
Hb_040963_010 |
0.1592248486 |
- |
- |
PREDICTED: polyneuridine-aldehyde esterase-like [Populus euphratica] |
| 6 |
Hb_148121_030 |
0.1820436442 |
- |
- |
PREDICTED: zinc-finger homeodomain protein 14 [Gossypium raimondii] |
| 7 |
Hb_079158_010 |
0.1842466385 |
- |
- |
PREDICTED: sperm acrosomal protein FSA-ACR.1-like [Jatropha curcas] |
| 8 |
Hb_101034_020 |
0.1925689079 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |
| 9 |
Hb_168509_020 |
0.1941055989 |
- |
- |
macrophage migration inhibitory factor family protein [Populus trichocarpa] |
| 10 |
Hb_001538_040 |
0.1949807176 |
- |
- |
PREDICTED: uncharacterized protein LOC104228304 [Nicotiana sylvestris] |
| 11 |
Hb_107809_010 |
0.1953916756 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 12 |
Hb_013786_010 |
0.1959565222 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_063523_010 |
0.1963840202 |
- |
- |
- |
| 14 |
Hb_007676_040 |
0.1974170838 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 15 |
Hb_002960_010 |
0.2014668043 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 16 |
Hb_000521_360 |
0.2061806271 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 17 |
Hb_001634_080 |
0.2069821771 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_011038_020 |
0.2173012184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005614_030 |
0.2185820277 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_005542_040 |
0.2185922341 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |