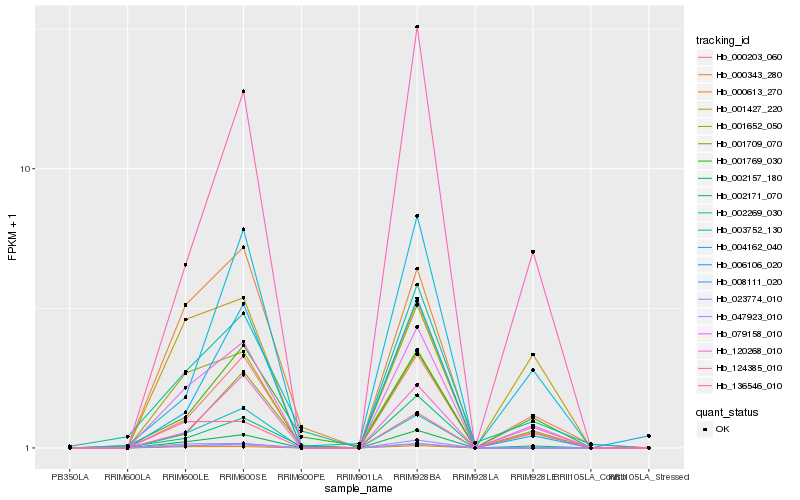
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079158_010 |
0.0 |
- |
- |
PREDICTED: sperm acrosomal protein FSA-ACR.1-like [Jatropha curcas] |
| 2 |
Hb_000203_060 |
0.1556208497 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0004s05660g [Populus trichocarpa] |
| 3 |
Hb_124385_010 |
0.1563646754 |
- |
- |
hypothetical protein JCGZ_13882 [Jatropha curcas] |
| 4 |
Hb_002157_180 |
0.1563733493 |
- |
- |
separase, putative [Ricinus communis] |
| 5 |
Hb_008111_020 |
0.1598866902 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 6 |
Hb_000343_280 |
0.1635654078 |
- |
- |
hypothetical protein JCGZ_24094 [Jatropha curcas] |
| 7 |
Hb_120268_010 |
0.1655252753 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1-like isoform X3 [Populus euphratica] |
| 8 |
Hb_002269_030 |
0.1677629821 |
- |
- |
hypothetical protein CISIN_1g018400mg [Citrus sinensis] |
| 9 |
Hb_001709_070 |
0.1734727131 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 10 |
Hb_047923_010 |
0.1750430317 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 11 |
Hb_003752_130 |
0.1752264774 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 12 |
Hb_001652_050 |
0.1753316473 |
- |
- |
- |
| 13 |
Hb_006106_020 |
0.1815120121 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 14 |
Hb_136546_010 |
0.1842466385 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1, partial [Populus euphratica] |
| 15 |
Hb_000613_270 |
0.1850251478 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 16 |
Hb_023774_010 |
0.1852560223 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 17 |
Hb_004162_040 |
0.1852690748 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Crassostrea gigas] |
| 18 |
Hb_002171_070 |
0.1859187509 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance gene-like protein ARGH30 [Populus trichocarpa] |
| 19 |
Hb_001769_030 |
0.1865238232 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 20 |
Hb_001427_220 |
0.1870645025 |
- |
- |
hypothetical protein CISIN_1g033375mg [Citrus sinensis] |