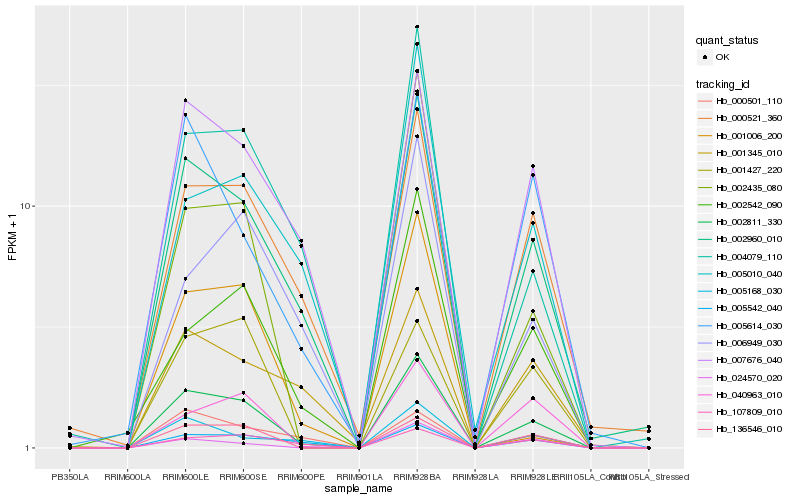
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_330 |
0.0 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 2 |
Hb_002960_010 |
0.1015042854 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 3 |
Hb_136546_010 |
0.1399717174 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1, partial [Populus euphratica] |
| 4 |
Hb_004079_110 |
0.1520083463 |
- |
- |
PREDICTED: uncharacterized protein LOC100257802 [Vitis vinifera] |
| 5 |
Hb_007676_040 |
0.153286946 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 6 |
Hb_000521_360 |
0.154064124 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 7 |
Hb_002435_080 |
0.1638916224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005542_040 |
0.1643702108 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 9 |
Hb_107809_010 |
0.1678861742 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 10 |
Hb_002542_090 |
0.1703765806 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 11 |
Hb_005168_030 |
0.1715014883 |
- |
- |
unknown [Zea mays] |
| 12 |
Hb_040963_010 |
0.1738138244 |
- |
- |
PREDICTED: polyneuridine-aldehyde esterase-like [Populus euphratica] |
| 13 |
Hb_005614_030 |
0.174238115 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005010_040 |
0.1799223925 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 15 |
Hb_024570_020 |
0.1824772787 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 16 |
Hb_006949_030 |
0.1853078943 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_001427_220 |
0.18849387 |
- |
- |
hypothetical protein CISIN_1g033375mg [Citrus sinensis] |
| 18 |
Hb_001345_010 |
0.1885199694 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |
| 19 |
Hb_000501_110 |
0.1914223153 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 20 |
Hb_001006_200 |
0.1924127521 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |