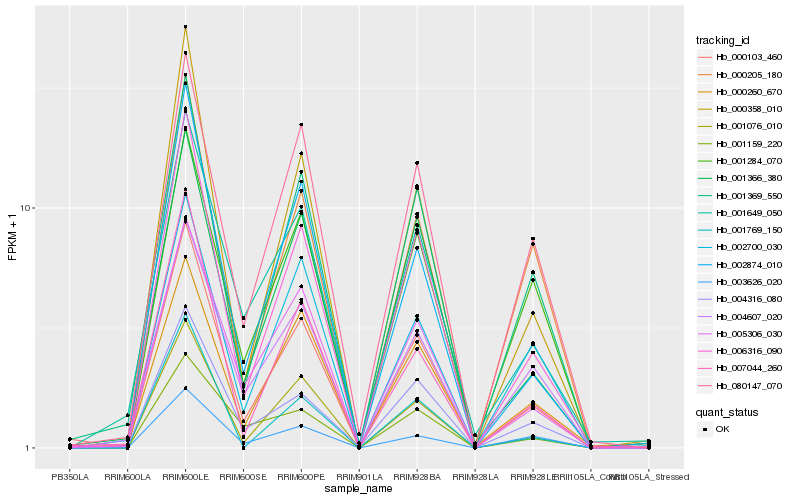
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_460 |
0.0 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 2 |
Hb_006316_090 |
0.0671767056 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_000358_010 |
0.0851081604 |
- |
- |
PREDICTED: basic blue protein [Populus euphratica] |
| 4 |
Hb_001076_010 |
0.0887396314 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_001369_550 |
0.0982584362 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 6 |
Hb_002874_010 |
0.1011602819 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |
| 7 |
Hb_005306_030 |
0.1061201409 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 8 |
Hb_001366_380 |
0.1077463564 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_007044_260 |
0.1089844577 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004316_080 |
0.1127917178 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 11 |
Hb_000205_180 |
0.1202292719 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 12 |
Hb_080147_070 |
0.1211651151 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 13 |
Hb_004607_020 |
0.1237727934 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 14 |
Hb_000260_670 |
0.1255162551 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 15 |
Hb_001159_220 |
0.126290328 |
- |
- |
ESC, putative [Ricinus communis] |
| 16 |
Hb_003626_020 |
0.1284465681 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 17 |
Hb_001649_050 |
0.1313911884 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 18 |
Hb_001769_150 |
0.1319112638 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 19 |
Hb_002700_030 |
0.1326568895 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 20 |
Hb_001284_070 |
0.1330740579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |