| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
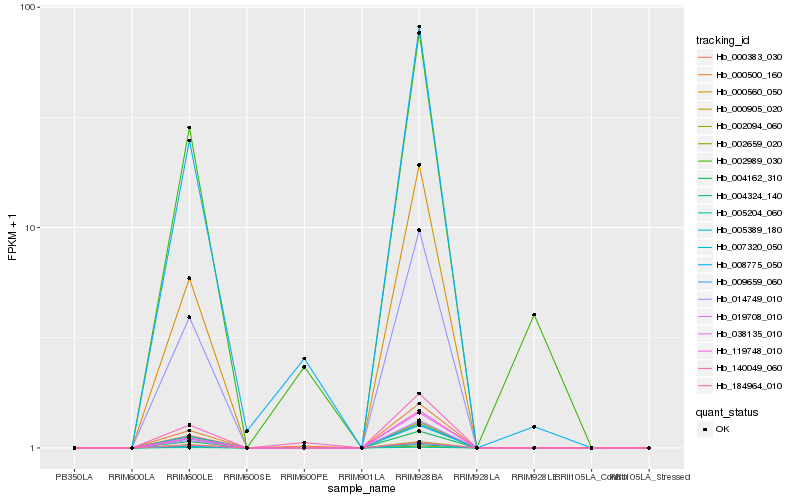
Hb_000500_160 |
0.0 |
- |
- |
hypothetical protein CISIN_1g039640mg, partial [Citrus sinensis] |
| 2 |
Hb_008775_050 |
0.0594325386 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 3 |
Hb_140049_060 |
0.0642138262 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 4 |
Hb_014749_010 |
0.1099132989 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 5 |
Hb_007320_050 |
0.1100108101 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 6 |
Hb_004162_310 |
0.1104881584 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 7 |
Hb_002659_020 |
0.1157610839 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 8 |
Hb_009659_060 |
0.116411482 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 9 |
Hb_000560_050 |
0.1176602878 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 10 |
Hb_038135_010 |
0.122672082 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 11 |
Hb_019708_010 |
0.1245100949 |
- |
- |
- |
| 12 |
Hb_002989_030 |
0.1250906779 |
- |
- |
PREDICTED: protein GAST1-like [Populus euphratica] |
| 13 |
Hb_005389_180 |
0.1281865942 |
- |
- |
PREDICTED: uncharacterized protein LOC105762407 [Gossypium raimondii] |
| 14 |
Hb_000383_030 |
0.1291883708 |
- |
- |
- |
| 15 |
Hb_119748_010 |
0.1339241781 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 16 |
Hb_000905_020 |
0.1344116297 |
- |
- |
PREDICTED: cytochrome P450 93A2-like [Jatropha curcas] |
| 17 |
Hb_004324_140 |
0.1362439129 |
- |
- |
PREDICTED: uncharacterized protein LOC105648358 [Jatropha curcas] |
| 18 |
Hb_184964_010 |
0.1366683654 |
- |
- |
- |
| 19 |
Hb_002094_060 |
0.1372396446 |
- |
- |
- |
| 20 |
Hb_005204_060 |
0.1379645026 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |