| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
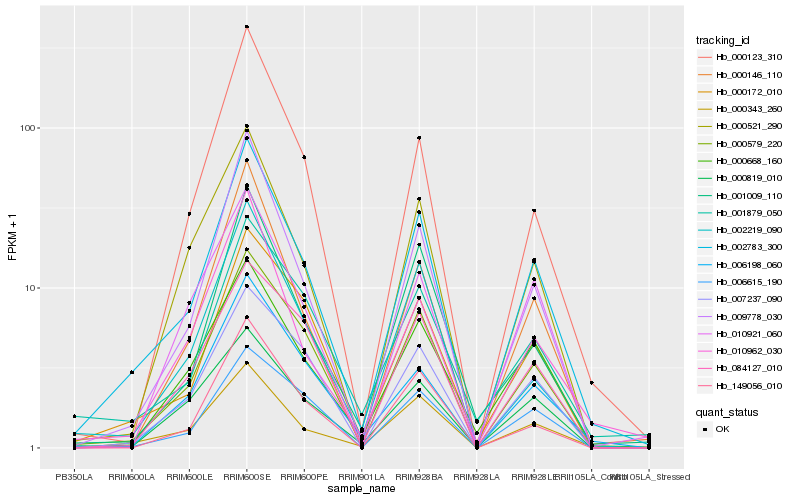
Hb_002783_300 |
0.0 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_000343_260 |
0.0851006652 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 3 |
Hb_000579_220 |
0.0992996051 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_009778_030 |
0.1079894267 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 5 |
Hb_000172_010 |
0.1192304415 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 6 |
Hb_000146_110 |
0.1237960679 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 7 |
Hb_000521_290 |
0.1240476908 |
- |
- |
hypothetical protein POPTR_0001s23530g [Populus trichocarpa] |
| 8 |
Hb_007237_090 |
0.1279313074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000668_160 |
0.1279578716 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 10 |
Hb_006198_060 |
0.1345358939 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 11 |
Hb_002219_090 |
0.1347212165 |
- |
- |
protein kinase atmrk1, putative [Ricinus communis] |
| 12 |
Hb_006615_190 |
0.1361309679 |
- |
- |
UDP-glycosyltransferase 1 [Linum usitatissimum] |
| 13 |
Hb_149056_010 |
0.1377816779 |
- |
- |
hypothetical protein CICLE_v10016664mg [Citrus clementina] |
| 14 |
Hb_001009_110 |
0.1380539605 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 15 |
Hb_000123_310 |
0.1399805129 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 16 |
Hb_010962_030 |
0.1414972145 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_001879_050 |
0.1435567331 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 18 |
Hb_010921_060 |
0.1436871793 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_000819_010 |
0.144156087 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 20 |
Hb_084127_010 |
0.1486284238 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |