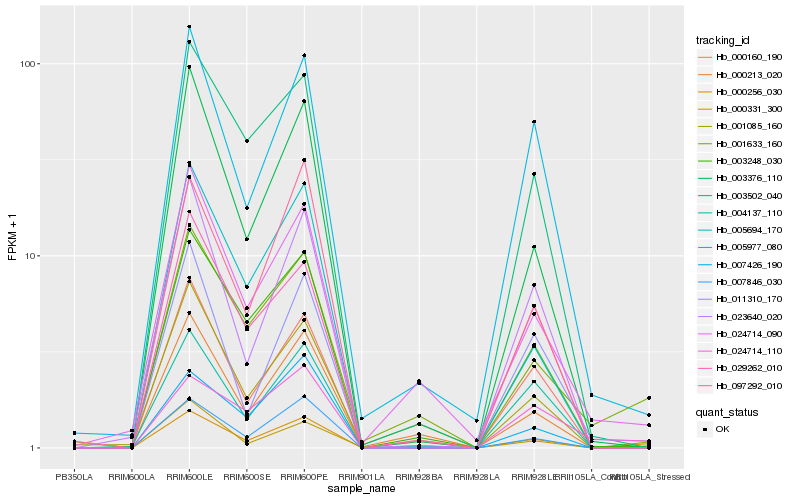
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_030 |
0.0 |
- |
- |
transferase, putative [Ricinus communis] |
| 2 |
Hb_001633_160 |
0.1475828117 |
- |
- |
PREDICTED: zerumbone synthase-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000331_300 |
0.1537470399 |
- |
- |
PREDICTED: malate synthase, glyoxysomal [Jatropha curcas] |
| 4 |
Hb_007846_030 |
0.161886594 |
- |
- |
Uncharacterized protein TCM_021839 [Theobroma cacao] |
| 5 |
Hb_003376_110 |
0.1631901227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000160_190 |
0.1654826681 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 7 |
Hb_003248_030 |
0.1655790898 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_000213_020 |
0.1685546138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005977_080 |
0.1711160458 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 10 |
Hb_005694_170 |
0.1734662264 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003502_040 |
0.175514597 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 12 |
Hb_023640_020 |
0.1755223261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_097292_010 |
0.176342721 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 14 |
Hb_011310_170 |
0.1770561699 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_024714_090 |
0.1797046716 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 16 |
Hb_001085_160 |
0.1834876562 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 17 |
Hb_004137_110 |
0.1835994381 |
- |
- |
PREDICTED: probable auxin efflux carrier component 6 [Jatropha curcas] |
| 18 |
Hb_007426_190 |
0.1858525241 |
- |
- |
PREDICTED: uncharacterized protein LOC103325200 [Prunus mume] |
| 19 |
Hb_029262_010 |
0.1864393215 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 20 |
Hb_024714_110 |
0.1876903164 |
- |
- |
PREDICTED: translocator protein homolog [Jatropha curcas] |