| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
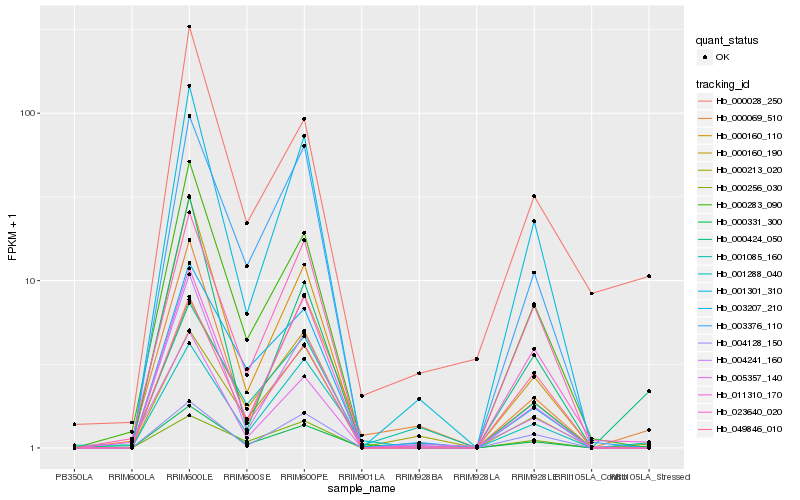
Hb_000331_300 |
0.0 |
- |
- |
PREDICTED: malate synthase, glyoxysomal [Jatropha curcas] |
| 2 |
Hb_000283_090 |
0.1253706366 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000069_510 |
0.1261517564 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 4 |
Hb_001301_310 |
0.1290573383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001288_040 |
0.1305265287 |
- |
- |
- |
| 6 |
Hb_003376_110 |
0.1367832959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000160_190 |
0.1488189202 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 8 |
Hb_000028_250 |
0.1499784988 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 9 |
Hb_000424_050 |
0.1507518245 |
- |
- |
erecta, putative [Ricinus communis] |
| 10 |
Hb_004241_160 |
0.1531368865 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-7 [Jatropha curcas] |
| 11 |
Hb_049846_010 |
0.1531534063 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 12 |
Hb_011310_170 |
0.1533179684 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000256_030 |
0.1537470399 |
- |
- |
transferase, putative [Ricinus communis] |
| 14 |
Hb_003207_210 |
0.1547225175 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 15 |
Hb_001085_160 |
0.154858644 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 16 |
Hb_023640_020 |
0.1559839795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004128_150 |
0.1586064296 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 18 |
Hb_000160_110 |
0.159415514 |
- |
- |
PREDICTED: uncharacterized protein LOC100253584 [Vitis vinifera] |
| 19 |
Hb_000213_020 |
0.1608572421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005357_140 |
0.1608593222 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |