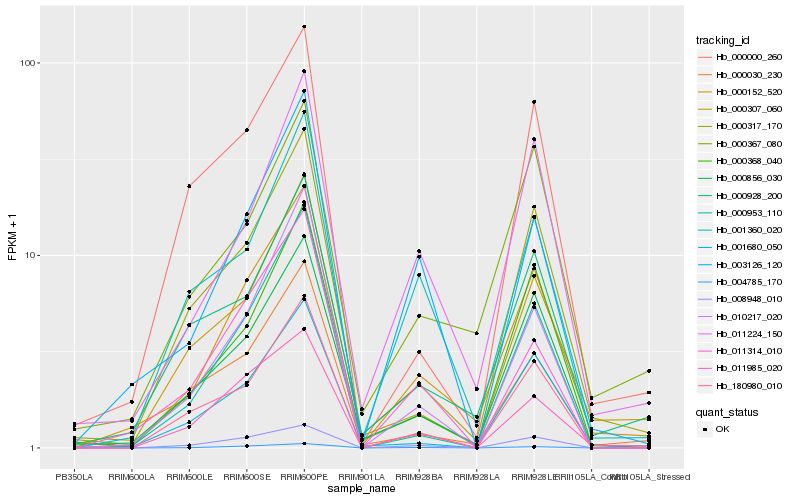
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_520 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 2 |
Hb_000856_030 |
0.119684939 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000317_170 |
0.1257665153 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 4 |
Hb_011224_150 |
0.1267490353 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000928_200 |
0.1348849219 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 6 |
Hb_003126_120 |
0.1377301017 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 7 |
Hb_011985_020 |
0.1417994249 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000367_080 |
0.1436240556 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 9 |
Hb_004785_170 |
0.1442443275 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_000368_040 |
0.144769338 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001360_020 |
0.1473075535 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 12 |
Hb_008948_010 |
0.1476493359 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 13 |
Hb_010217_020 |
0.1497411579 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 14 |
Hb_001680_050 |
0.1506580996 |
- |
- |
- |
| 15 |
Hb_000953_110 |
0.1507382203 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 16 |
Hb_000307_060 |
0.1509199901 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 17 |
Hb_180980_010 |
0.1529828609 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 18 |
Hb_000030_230 |
0.153417669 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 19 |
Hb_011314_010 |
0.1552847011 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000000_260 |
0.1589053638 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |