| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
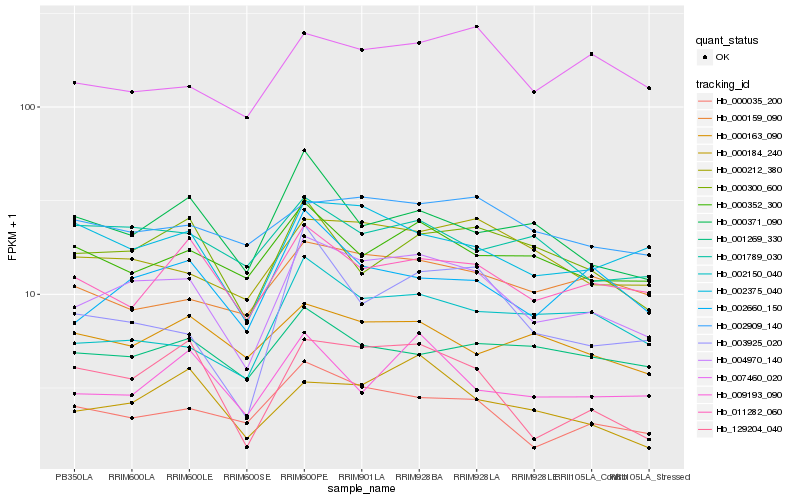
Hb_004970_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633635 [Jatropha curcas] |
| 2 |
Hb_000035_200 |
0.1139689619 |
- |
- |
PREDICTED: phospholipase D Z-like [Jatropha curcas] |
| 3 |
Hb_000212_380 |
0.114611406 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002375_040 |
0.1180443478 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 5 |
Hb_002660_150 |
0.1196340151 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011282_060 |
0.122615873 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 7 |
Hb_000184_240 |
0.1237064446 |
- |
- |
PREDICTED: uncharacterized protein LOC105647985 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000300_600 |
0.1239511642 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 9 |
Hb_002150_040 |
0.1246870887 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 10 |
Hb_007460_020 |
0.1297568756 |
- |
- |
PREDICTED: guanosine nucleotide diphosphate dissociation inhibitor 1 [Jatropha curcas] |
| 11 |
Hb_000352_300 |
0.1299921474 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 12 |
Hb_002909_140 |
0.1304644301 |
- |
- |
PREDICTED: D-xylose-proton symporter-like 2 [Jatropha curcas] |
| 13 |
Hb_129204_040 |
0.1306439873 |
- |
- |
PREDICTED: meiotic nuclear division protein 1 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_000159_090 |
0.1312024188 |
- |
- |
hypothetical protein POPTR_0015s153201g, partial [Populus trichocarpa] |
| 15 |
Hb_000371_090 |
0.1322957973 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 16 |
Hb_001789_030 |
0.1346331124 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 17 |
Hb_001269_330 |
0.1349254607 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 18 |
Hb_003925_020 |
0.1358599891 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 19 |
Hb_000163_090 |
0.137205813 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 20 |
Hb_009193_090 |
0.1373948026 |
- |
- |
PREDICTED: protein root UVB sensitive 3 isoform X1 [Jatropha curcas] |