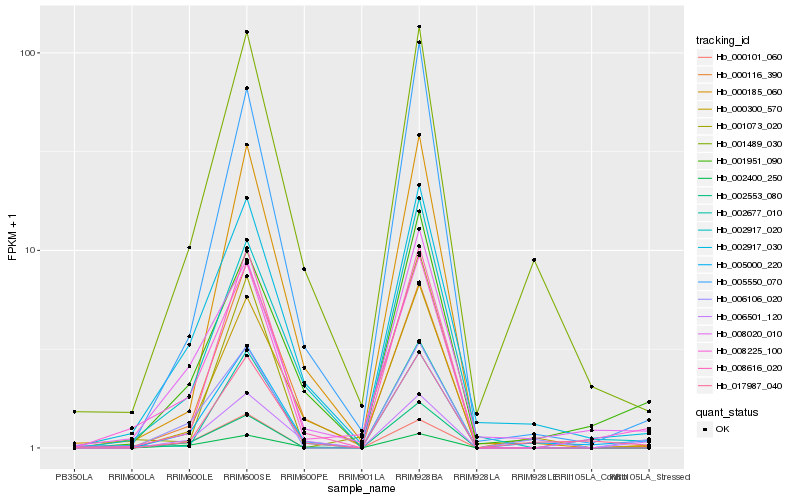
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008225_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_008020_010 |
0.1267937184 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like [Jatropha curcas] |
| 3 |
Hb_002917_030 |
0.1449040192 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Setaria italica] |
| 4 |
Hb_005000_220 |
0.1496110846 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 5 |
Hb_001951_090 |
0.1677526998 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_006106_020 |
0.1720917341 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 7 |
Hb_002553_080 |
0.1721051881 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF096-like [Eucalyptus grandis] |
| 8 |
Hb_017987_040 |
0.1727332015 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 9 |
Hb_006501_120 |
0.1748514989 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 10 |
Hb_002917_020 |
0.175294119 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 11 |
Hb_001489_030 |
0.1754301089 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 12 |
Hb_001073_020 |
0.1772272797 |
- |
- |
hypothetical protein CISIN_1g031854mg [Citrus sinensis] |
| 13 |
Hb_008616_020 |
0.1778874873 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000101_060 |
0.1780916512 |
- |
- |
- |
| 15 |
Hb_000116_390 |
0.1782213781 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 16 |
Hb_005550_070 |
0.1785596521 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 17 |
Hb_002400_250 |
0.1801975625 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000300_570 |
0.1816433444 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 19 |
Hb_002677_010 |
0.1824870732 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 20 |
Hb_000185_060 |
0.1825745652 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |