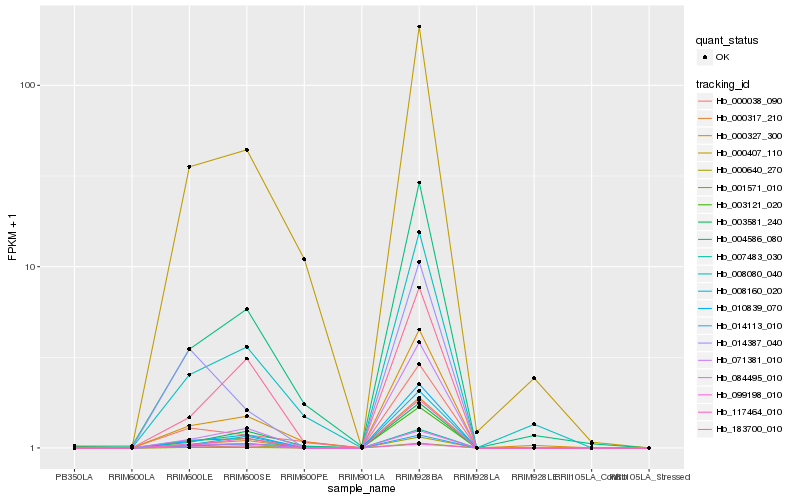
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003121_020 |
0.0 |
- |
- |
secretory peroxidase PX3 [Manihot esculenta] |
| 2 |
Hb_084495_010 |
0.0345943299 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Eucalyptus grandis] |
| 3 |
Hb_099198_010 |
0.0517085111 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At2g25790 isoform X2 [Nicotiana tomentosiformis] |
| 4 |
Hb_014113_010 |
0.08599066 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_010839_070 |
0.0914693706 |
- |
- |
uncharacterized protein LOC100810725 [Glycine max] |
| 6 |
Hb_001571_010 |
0.1116730654 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 7 |
Hb_000640_270 |
0.1139136822 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable pectinesterase 30 [Jatropha curcas] |
| 8 |
Hb_000327_300 |
0.1305660546 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 9 |
Hb_117464_010 |
0.1339564955 |
- |
- |
PREDICTED: flavone 3'-O-methyltransferase 1-like [Jatropha curcas] |
| 10 |
Hb_004586_080 |
0.1352215677 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 11 |
Hb_008160_020 |
0.1398422018 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 23-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_183700_010 |
0.1416091428 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000407_110 |
0.1419708526 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 14 |
Hb_003581_240 |
0.1462682454 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 15 |
Hb_071381_010 |
0.1528084032 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 16 |
Hb_008080_040 |
0.1544399159 |
- |
- |
hypothetical protein EUGRSUZ_D00834 [Eucalyptus grandis] |
| 17 |
Hb_000038_090 |
0.1582880648 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |
| 18 |
Hb_014387_040 |
0.1592705056 |
- |
- |
PREDICTED: uncharacterized protein LOC105636953 [Jatropha curcas] |
| 19 |
Hb_000317_210 |
0.1593859579 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39 [Jatropha curcas] |
| 20 |
Hb_007483_030 |
0.159454275 |
- |
- |
- |