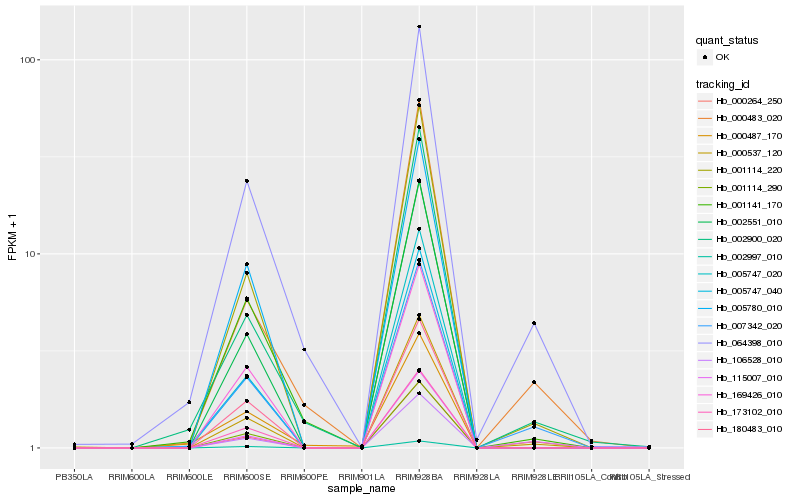
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007342_020 |
0.0 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_115007_010 |
0.0741656149 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 3 |
Hb_000537_120 |
0.0876900023 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_064398_010 |
0.0888337715 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_001114_220 |
0.0941837804 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 6 |
Hb_000487_170 |
0.108947896 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000483_020 |
0.1200077149 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_001114_290 |
0.1245619785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002900_020 |
0.1246942115 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 10 |
Hb_173102_010 |
0.1257415418 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_005747_040 |
0.1261280363 |
- |
- |
PREDICTED: peroxidase N1-like, partial [Nicotiana sylvestris] |
| 12 |
Hb_106528_010 |
0.126372763 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_001141_170 |
0.1278216711 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 14 |
Hb_002551_010 |
0.1283438191 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_002997_010 |
0.1292687696 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Malus domestica] |
| 16 |
Hb_005747_020 |
0.1306054805 |
- |
- |
bacterial-induced peroxidase [Gossypium schwendimanii] |
| 17 |
Hb_169426_010 |
0.1311655616 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_005780_010 |
0.1312566291 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 19 |
Hb_000264_250 |
0.1313587912 |
- |
- |
thioredoxin family protein [Arabidopsis thaliana] |
| 20 |
Hb_180483_010 |
0.1314403207 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |