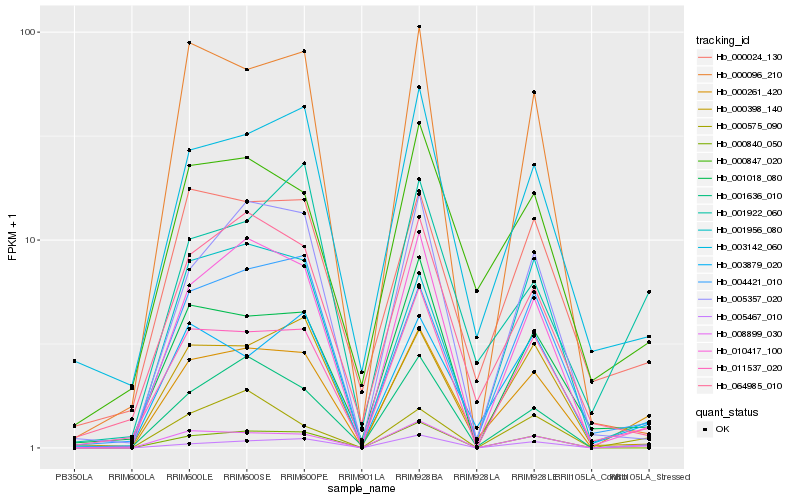
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_420 |
0.0 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 2 |
Hb_008899_030 |
0.1130775056 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 3 |
Hb_010417_100 |
0.1350266963 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 4 |
Hb_005467_010 |
0.1360075451 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011537_020 |
0.1426496191 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 6 |
Hb_003879_020 |
0.1482604343 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 7 |
Hb_003142_060 |
0.1544785317 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000024_130 |
0.1587170487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000847_020 |
0.1599834602 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 10 |
Hb_001636_010 |
0.1639518549 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 11 |
Hb_004421_010 |
0.1640867092 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_001018_080 |
0.1652822255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000398_140 |
0.168484332 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 14 |
Hb_001956_080 |
0.1691351796 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 15 |
Hb_001922_060 |
0.1702086856 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000840_050 |
0.1706753696 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 17 |
Hb_005357_020 |
0.1708961648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_064985_010 |
0.174075308 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 19 |
Hb_000575_090 |
0.1766924564 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Populus euphratica] |
| 20 |
Hb_000096_210 |
0.1767030273 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |