| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
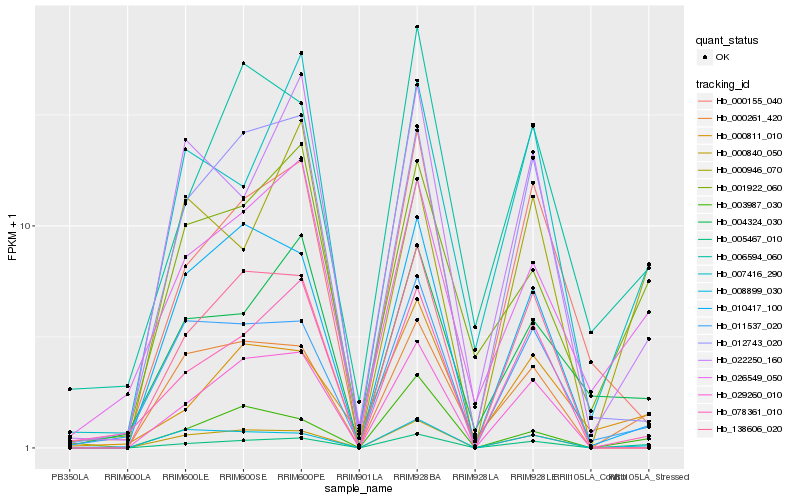
Hb_005467_010 |
0.0 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000261_420 |
0.1360075451 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 3 |
Hb_008899_030 |
0.167606461 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 4 |
Hb_003987_030 |
0.1719246041 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 5 |
Hb_011537_020 |
0.1809892394 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 6 |
Hb_026549_050 |
0.1828560573 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001922_060 |
0.186583654 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_078361_010 |
0.1885674626 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 9 |
Hb_000811_010 |
0.1897669741 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 10 |
Hb_007416_290 |
0.190479815 |
- |
- |
PREDICTED: plasma membrane ATPase 4-like [Pyrus x bretschneideri] |
| 11 |
Hb_010417_100 |
0.1913028215 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 12 |
Hb_000946_070 |
0.1959580528 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 13 |
Hb_006594_060 |
0.198135466 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 14 |
Hb_138606_020 |
0.1984294533 |
- |
- |
Nitrate transporter2.5 [Theobroma cacao] |
| 15 |
Hb_029260_010 |
0.1993521603 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 16 |
Hb_012743_020 |
0.2025864933 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_000840_050 |
0.2027867139 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 18 |
Hb_004324_030 |
0.203478889 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 19 |
Hb_000155_040 |
0.2036469852 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 20 |
Hb_022250_160 |
0.2036978107 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |