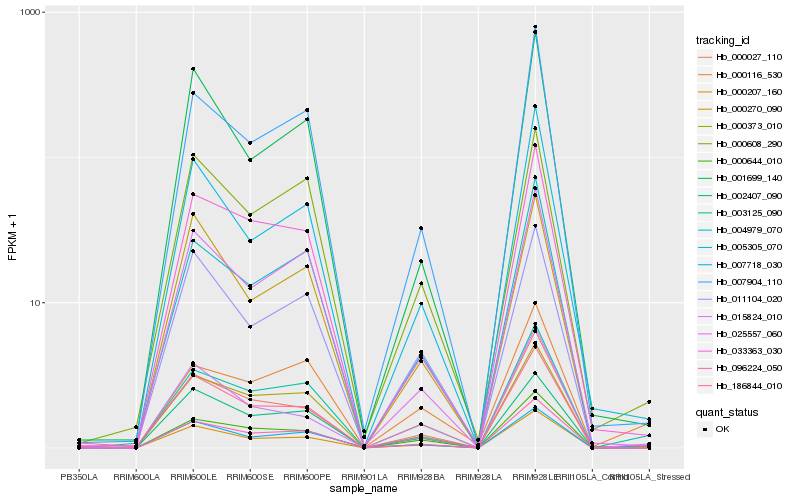
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000207_160 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Populus euphratica] |
| 2 |
Hb_003125_090 |
0.0810430095 |
- |
- |
- |
| 3 |
Hb_015824_010 |
0.1099751293 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 4 |
Hb_096224_050 |
0.1178111338 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 5 |
Hb_000373_010 |
0.1182658911 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_005305_070 |
0.1206275973 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 7 |
Hb_000027_110 |
0.1282431549 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 8 |
Hb_001699_140 |
0.1298744442 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 9 |
Hb_033363_030 |
0.1310489242 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000644_010 |
0.1316816967 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002407_090 |
0.1321780704 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 12 |
Hb_000608_290 |
0.1322380347 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 13 |
Hb_000270_090 |
0.1326449861 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 14 |
Hb_011104_020 |
0.1328804408 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 15 |
Hb_007904_110 |
0.1336533174 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_004979_070 |
0.1366301871 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 17 |
Hb_007718_030 |
0.1374563853 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_000116_530 |
0.1378131623 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 3; Short=HMG-CoA reductase 3 [Hevea brasiliensis] |
| 19 |
Hb_186844_010 |
0.1378497835 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_025557_060 |
0.1387227711 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |