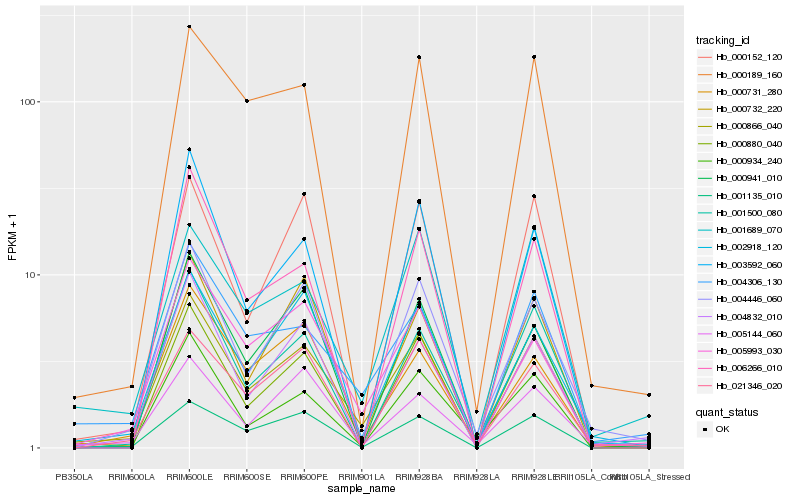
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_040 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 2 |
Hb_004306_130 |
0.1011847035 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005993_030 |
0.1149469182 |
- |
- |
putative senescence-associated protein SAG102 [Populus trichocarpa] |
| 4 |
Hb_001135_010 |
0.1178196061 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 5 |
Hb_002918_120 |
0.1200220233 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_004446_060 |
0.1239557532 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 7 |
Hb_001500_080 |
0.1375463443 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000189_160 |
0.1387878673 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 9 |
Hb_000732_220 |
0.1393680879 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 10 |
Hb_001689_070 |
0.13982993 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_004832_010 |
0.1413698812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000731_280 |
0.1443873306 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000880_040 |
0.144854034 |
- |
- |
- |
| 14 |
Hb_000934_240 |
0.1473521732 |
- |
- |
PREDICTED: peroxidase 13 [Jatropha curcas] |
| 15 |
Hb_003592_060 |
0.1485863631 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |
| 16 |
Hb_005144_060 |
0.14868959 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 17 |
Hb_000941_010 |
0.1486929146 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 18 |
Hb_006266_010 |
0.1490926665 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_021346_020 |
0.1502316721 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 20 |
Hb_000152_120 |
0.1510860932 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |