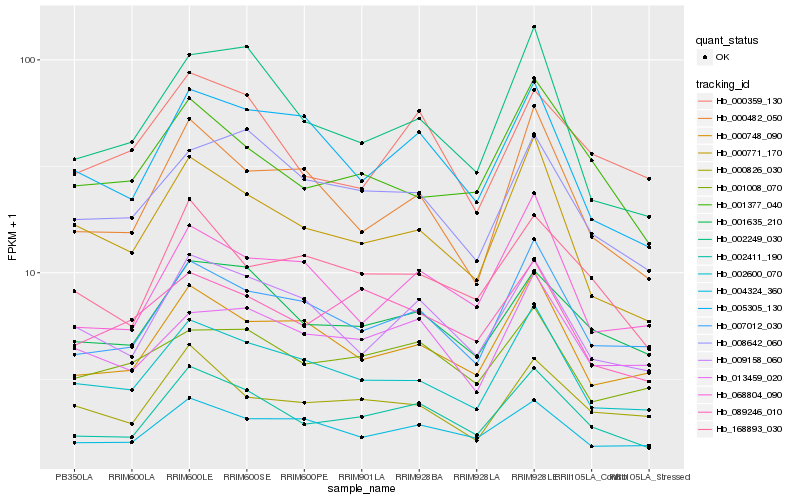
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002411_190 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 2 |
Hb_001635_210 |
0.0691096728 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007012_030 |
0.0810714048 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002600_070 |
0.09027229 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 5 |
Hb_009158_060 |
0.090629351 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 6 |
Hb_168893_030 |
0.0919539175 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 7 |
Hb_000748_090 |
0.0963949637 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 8 |
Hb_004324_360 |
0.0968541804 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000359_130 |
0.0983180742 |
- |
- |
4-hydroxyphenylpyruvate dioxygenase [Hevea brasiliensis] |
| 10 |
Hb_000826_030 |
0.1037372435 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 11 |
Hb_001377_040 |
0.1038001937 |
- |
- |
chitinase, putative [Ricinus communis] |
| 12 |
Hb_008642_060 |
0.1040179117 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 13 |
Hb_000771_170 |
0.1040180523 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 14 |
Hb_089246_010 |
0.1043901209 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 15 |
Hb_005305_130 |
0.1048697482 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 16 |
Hb_001008_070 |
0.1064821746 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 17 |
Hb_068804_090 |
0.1069008231 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 18 |
Hb_000482_050 |
0.1071211994 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_013459_020 |
0.1078479398 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 20 |
Hb_002249_030 |
0.1081211836 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |