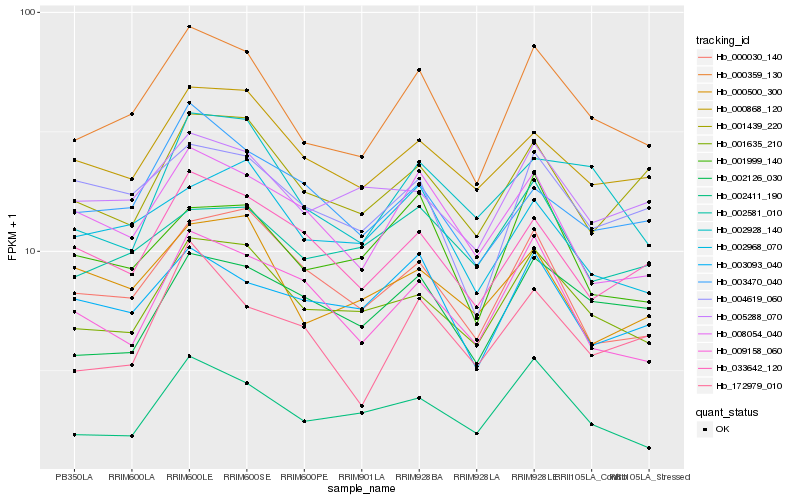
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_130 |
0.0 |
- |
- |
4-hydroxyphenylpyruvate dioxygenase [Hevea brasiliensis] |
| 2 |
Hb_001635_210 |
0.0779383559 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002411_190 |
0.0983180742 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 4 |
Hb_000868_120 |
0.0994680404 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 5 |
Hb_005288_070 |
0.1012684706 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_002126_030 |
0.1015110032 |
- |
- |
ferrochelatase, putative [Ricinus communis] |
| 7 |
Hb_002928_140 |
0.1034882113 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004619_060 |
0.103807053 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 9 |
Hb_001999_140 |
0.1044433132 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_001439_220 |
0.1046467484 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 11 |
Hb_033642_120 |
0.1060829694 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 12 |
Hb_008054_040 |
0.1061818241 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 13 |
Hb_003093_040 |
0.1070980429 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_000030_140 |
0.1073847191 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002968_070 |
0.1089571099 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 16 |
Hb_003470_040 |
0.109651973 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_002581_010 |
0.1103849181 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 18 |
Hb_009158_060 |
0.111604564 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 19 |
Hb_172979_010 |
0.1116352717 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 20 |
Hb_000500_300 |
0.1156550289 |
- |
- |
PREDICTED: nucleolar protein 10 [Jatropha curcas] |